Probe CUST_19050_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19050_PI426222305 | JHI_St_60k_v1 | DMT400052620 | GTTATAAAGTGTTTATTGCAGCATTACTCAATGTGTCTGTGCAGTGGACAACTGGTATCT |
All Microarray Probes Designed to Gene DMG400020426
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19042_PI426222305 | JHI_St_60k_v1 | DMT400052621 | GCAGAAGAACATGGAAATACTCAAATTTCAACTGTCATTTTTGATTTGGATGGTACCCTT |
| CUST_19050_PI426222305 | JHI_St_60k_v1 | DMT400052620 | GTTATAAAGTGTTTATTGCAGCATTACTCAATGTGTCTGTGCAGTGGACAACTGGTATCT |
Microarray Signals from CUST_19050_PI426222305
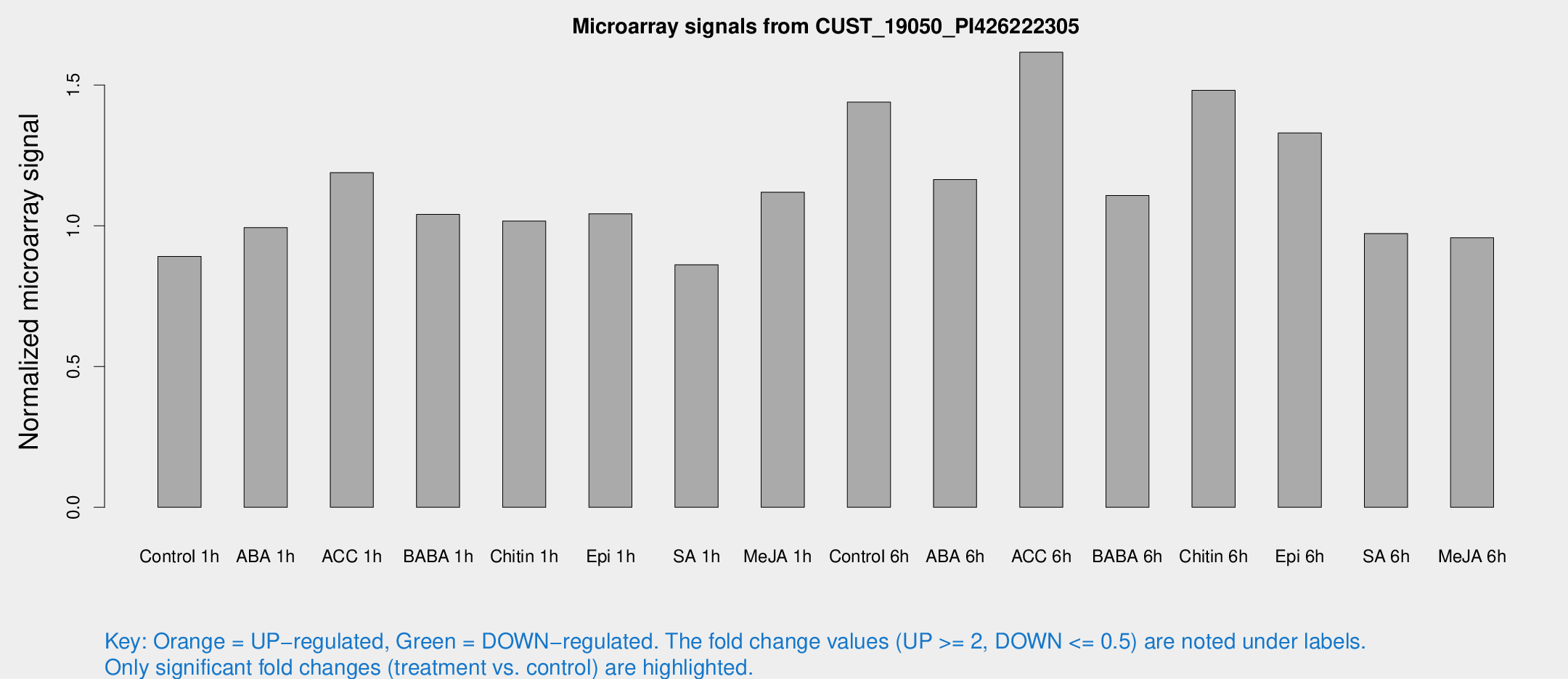
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.01094 | 2.90578 | 0.891316 | 0.510358 |
| ABA 1h | 4.81509 | 2.79251 | 0.993384 | 0.554317 |
| ACC 1h | 7.29728 | 3.18201 | 1.18881 | 0.578426 |
| BABA 1h | 5.72467 | 3.08491 | 1.04037 | 0.565017 |
| Chitin 1h | 5.0211 | 2.92216 | 1.01706 | 0.567569 |
| Epi 1h | 5.02514 | 2.91672 | 1.04274 | 0.590554 |
| SA 1h | 5.03452 | 2.91666 | 0.861305 | 0.498973 |
| Me-JA 1h | 5.11406 | 2.96895 | 1.11898 | 0.636041 |
| Control 6h | 10.2161 | 4.98045 | 1.43935 | 0.70477 |
| ABA 6h | 7.46203 | 3.15404 | 1.16446 | 0.561463 |
| ACC 6h | 10.6546 | 3.70449 | 1.61665 | 0.558918 |
| BABA 6h | 7.49184 | 3.39347 | 1.10754 | 0.552111 |
| Chitin 6h | 9.48866 | 3.361 | 1.4816 | 0.594205 |
| Epi 6h | 9.46049 | 3.51465 | 1.33015 | 0.621719 |
| SA 6h | 5.47621 | 3.17298 | 0.972421 | 0.563111 |
| Me-JA 6h | 5.40583 | 3.0045 | 0.957649 | 0.530208 |
Source Transcript PGSC0003DMT400052620 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G21470.1 | +1 | 3e-60 | 200 | 127/270 (47%) | riboflavin kinase/FMN hydrolase | chr4:11431284-11433197 FORWARD LENGTH=379 |
