Probe CUST_19026_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19026_PI426222305 | JHI_St_60k_v1 | DMT400044674 | GGATAAGCCAAATTAGTGTTAAGAGTTCTCTGTATTCCTTCATTTCTCTTCTTCTCTGTA |
All Microarray Probes Designed to Gene DMG400017331
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19026_PI426222305 | JHI_St_60k_v1 | DMT400044674 | GGATAAGCCAAATTAGTGTTAAGAGTTCTCTGTATTCCTTCATTTCTCTTCTTCTCTGTA |
Microarray Signals from CUST_19026_PI426222305
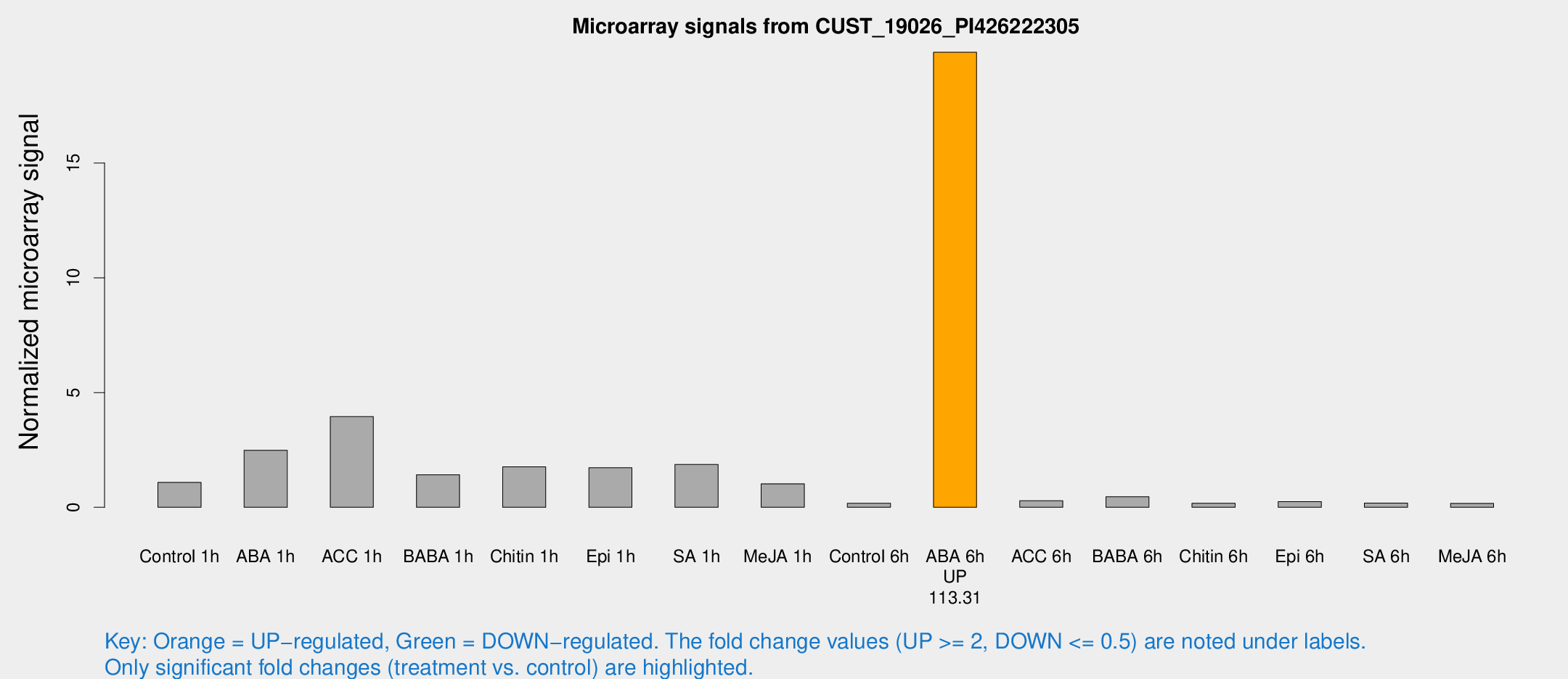
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 43.491 | 16.78 | 1.08154 | 0.344617 |
| ABA 1h | 84.0916 | 24.5509 | 2.48745 | 0.897647 |
| ACC 1h | 145.604 | 21.4753 | 3.95213 | 0.896146 |
| BABA 1h | 49.1813 | 7.06419 | 1.41812 | 0.133538 |
| Chitin 1h | 65.0496 | 26.7067 | 1.7655 | 0.780517 |
| Epi 1h | 52.9148 | 5.34755 | 1.72169 | 0.191203 |
| SA 1h | 76.0837 | 22.6671 | 1.86741 | 0.63366 |
| Me-JA 1h | 35.4637 | 12.5704 | 1.02318 | 0.485071 |
| Control 6h | 6.17885 | 3.54234 | 0.175014 | 0.10029 |
| ABA 6h | 858.948 | 337.339 | 19.8313 | 6.92358 |
| ACC 6h | 12.2604 | 4.20829 | 0.283187 | 0.110771 |
| BABA 6h | 24.7952 | 12.4825 | 0.460007 | 0.355884 |
| Chitin 6h | 6.70964 | 3.89279 | 0.179172 | 0.103891 |
| Epi 6h | 11.3101 | 4.33809 | 0.248035 | 0.115447 |
| SA 6h | 6.33069 | 3.67126 | 0.182036 | 0.105557 |
| Me-JA 6h | 6.01661 | 3.49161 | 0.17154 | 0.0993533 |
Source Transcript PGSC0003DMT400044674 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G78830.1 | +3 | 3e-34 | 134 | 111/342 (32%) | Curculin-like (mannose-binding) lectin family protein | chr1:29637141-29638508 REVERSE LENGTH=455 |
