Probe CUST_18866_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18866_PI426222305 | JHI_St_60k_v1 | DMT400001119 | ACCGTGTGTGGCATTAGATATGGTTGCTAGCATCACACAATCTCCAGCAGTTTCAAAAAA |
All Microarray Probes Designed to Gene DMG400000422
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18783_PI426222305 | JHI_St_60k_v1 | DMT400001118 | ACCGTGTGTGGCATTAGATATGGTTGCTAGCATCACACAATCTCCAGCAGTTTCAAAAAA |
| CUST_18866_PI426222305 | JHI_St_60k_v1 | DMT400001119 | ACCGTGTGTGGCATTAGATATGGTTGCTAGCATCACACAATCTCCAGCAGTTTCAAAAAA |
Microarray Signals from CUST_18866_PI426222305
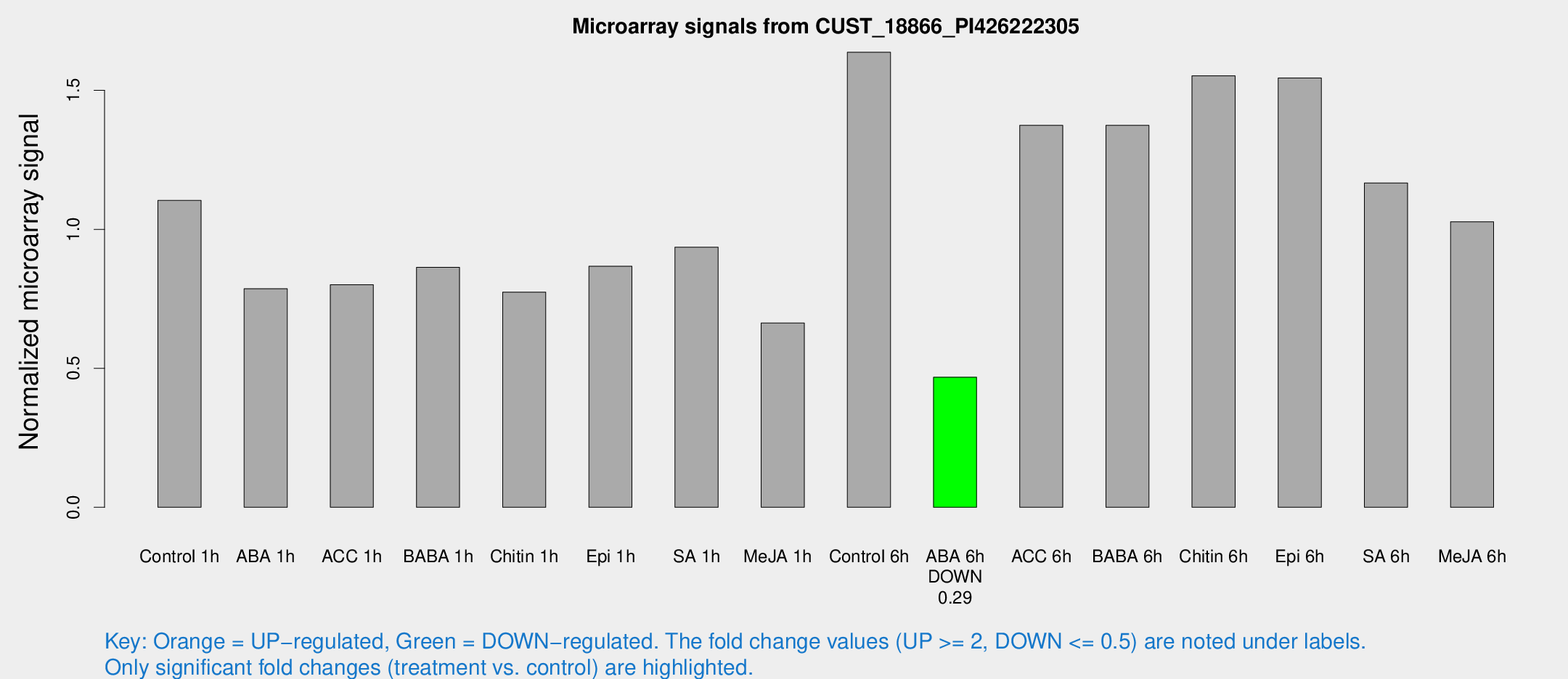
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2513.28 | 426.332 | 1.104 | 0.11971 |
| ABA 1h | 1559.71 | 162.318 | 0.786235 | 0.0880691 |
| ACC 1h | 2062.01 | 611.239 | 0.800705 | 0.256537 |
| BABA 1h | 1880.42 | 253.223 | 0.863251 | 0.0498646 |
| Chitin 1h | 1547.1 | 89.5927 | 0.774213 | 0.0864142 |
| Epi 1h | 1670.81 | 108.852 | 0.866946 | 0.0525479 |
| SA 1h | 2157.11 | 246.464 | 0.935558 | 0.108916 |
| Me-JA 1h | 1202.85 | 69.7034 | 0.662777 | 0.0383056 |
| Control 6h | 3851.73 | 843.515 | 1.63709 | 0.270845 |
| ABA 6h | 1104.63 | 63.9483 | 0.468014 | 0.0352912 |
| ACC 6h | 3528.35 | 343.009 | 1.37391 | 0.0961998 |
| BABA 6h | 3468.28 | 466.258 | 1.37409 | 0.182895 |
| Chitin 6h | 3667.03 | 211.969 | 1.55229 | 0.089634 |
| Epi 6h | 3918.66 | 487.2 | 1.54441 | 0.247702 |
| SA 6h | 2565.16 | 148.529 | 1.16677 | 0.131965 |
| Me-JA 6h | 2357.22 | 443.524 | 1.0274 | 0.119237 |
Source Transcript PGSC0003DMT400001119 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G27920.1 | +3 | 0.0 | 598 | 298/440 (68%) | serine carboxypeptidase-like 51 | chr2:11885777-11889043 REVERSE LENGTH=461 |
