Probe CUST_18274_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18274_PI426222305 | JHI_St_60k_v1 | DMT400042158 | ATTGGATCTCTCATCCAACAAAATCAGCGGAGAAATTCCACAACAACTTGCATCCCTCAA |
All Microarray Probes Designed to Gene DMG400016357
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18274_PI426222305 | JHI_St_60k_v1 | DMT400042158 | ATTGGATCTCTCATCCAACAAAATCAGCGGAGAAATTCCACAACAACTTGCATCCCTCAA |
| CUST_18302_PI426222305 | JHI_St_60k_v1 | DMT400042159 | AATGAGAAGGCACAAGAAAAGACATTAGTGTGTAACCTCGAGGTATCAACTTAATCTCTA |
Microarray Signals from CUST_18274_PI426222305
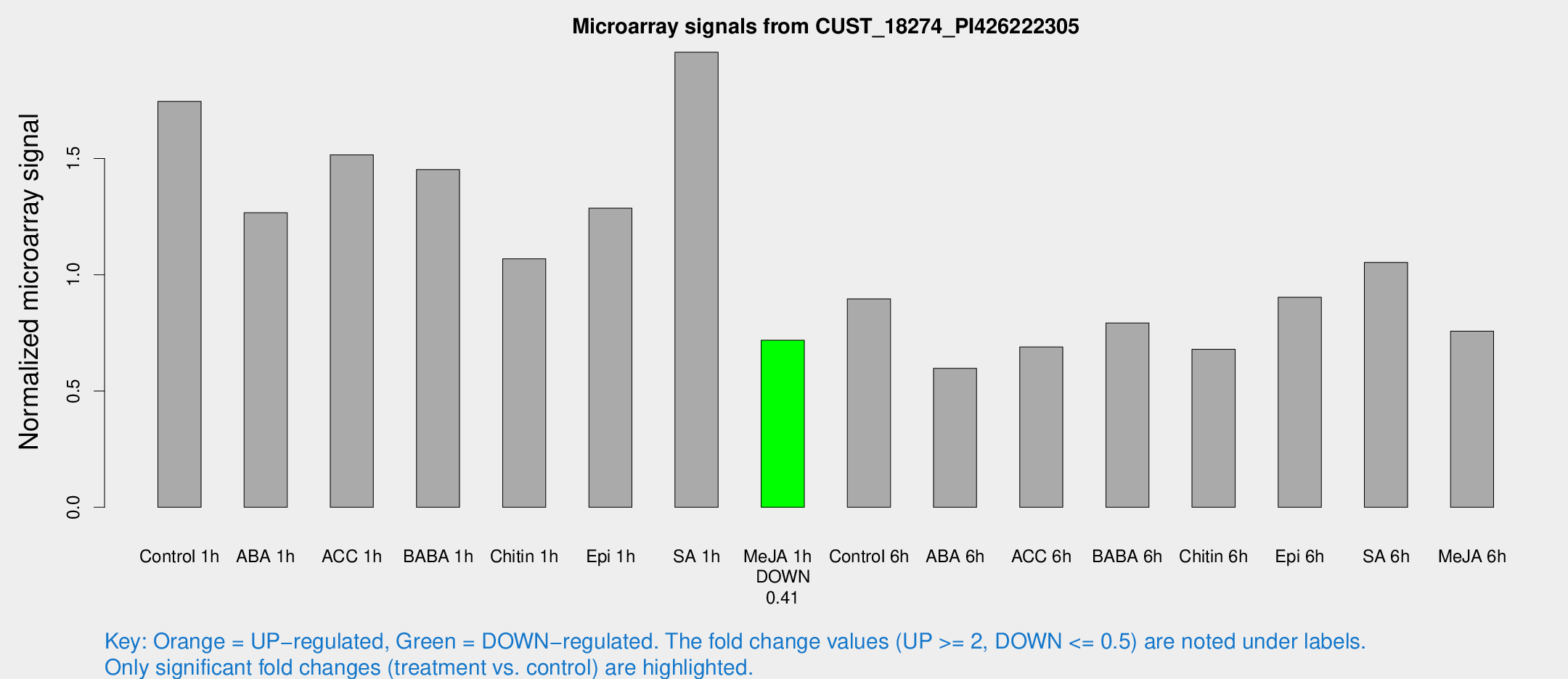
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5351.29 | 1046.98 | 1.74541 | 0.216831 |
| ABA 1h | 3366.02 | 397.601 | 1.26668 | 0.0887436 |
| ACC 1h | 4981.52 | 1232.23 | 1.5158 | 0.340273 |
| BABA 1h | 4463.72 | 1083.27 | 1.45254 | 0.27288 |
| Chitin 1h | 2878.89 | 329.337 | 1.06884 | 0.0617216 |
| Epi 1h | 3313.77 | 253.668 | 1.28615 | 0.0907359 |
| SA 1h | 6022.09 | 686.075 | 1.95684 | 0.112983 |
| Me-JA 1h | 1794.2 | 342.241 | 0.718451 | 0.0793552 |
| Control 6h | 2828.46 | 641.404 | 0.896048 | 0.15875 |
| ABA 6h | 2025.52 | 519.141 | 0.597717 | 0.141494 |
| ACC 6h | 2345.63 | 135.811 | 0.689393 | 0.0827651 |
| BABA 6h | 2674.07 | 342.354 | 0.79288 | 0.0817893 |
| Chitin 6h | 2154.69 | 173.095 | 0.679663 | 0.0506946 |
| Epi 6h | 3126.94 | 609.257 | 0.902976 | 0.153371 |
| SA 6h | 3202.46 | 659.48 | 1.05272 | 0.176794 |
| Me-JA 6h | 2437.31 | 704.121 | 0.757572 | 0.169229 |
Source Transcript PGSC0003DMT400042158 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G47890.1 | +1 | 2e-59 | 211 | 148/414 (36%) | receptor like protein 7 | chr1:17643976-17647035 FORWARD LENGTH=1019 |
