Probe CUST_18141_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18141_PI426222305 | JHI_St_60k_v1 | DMT400048452 | CTTGGCTTCTCCTTTTTTAATTGGAGTAAGTCGAAATGGTTCTCTGATATTAGTAAGATG |
All Microarray Probes Designed to Gene DMG400018819
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18141_PI426222305 | JHI_St_60k_v1 | DMT400048452 | CTTGGCTTCTCCTTTTTTAATTGGAGTAAGTCGAAATGGTTCTCTGATATTAGTAAGATG |
| CUST_18167_PI426222305 | JHI_St_60k_v1 | DMT400048453 | GGATGTCTTCAAGATACTAGTCTTGAATCTTGATCTTCTTAGAAATGTGAAGCAGTGATT |
Microarray Signals from CUST_18141_PI426222305
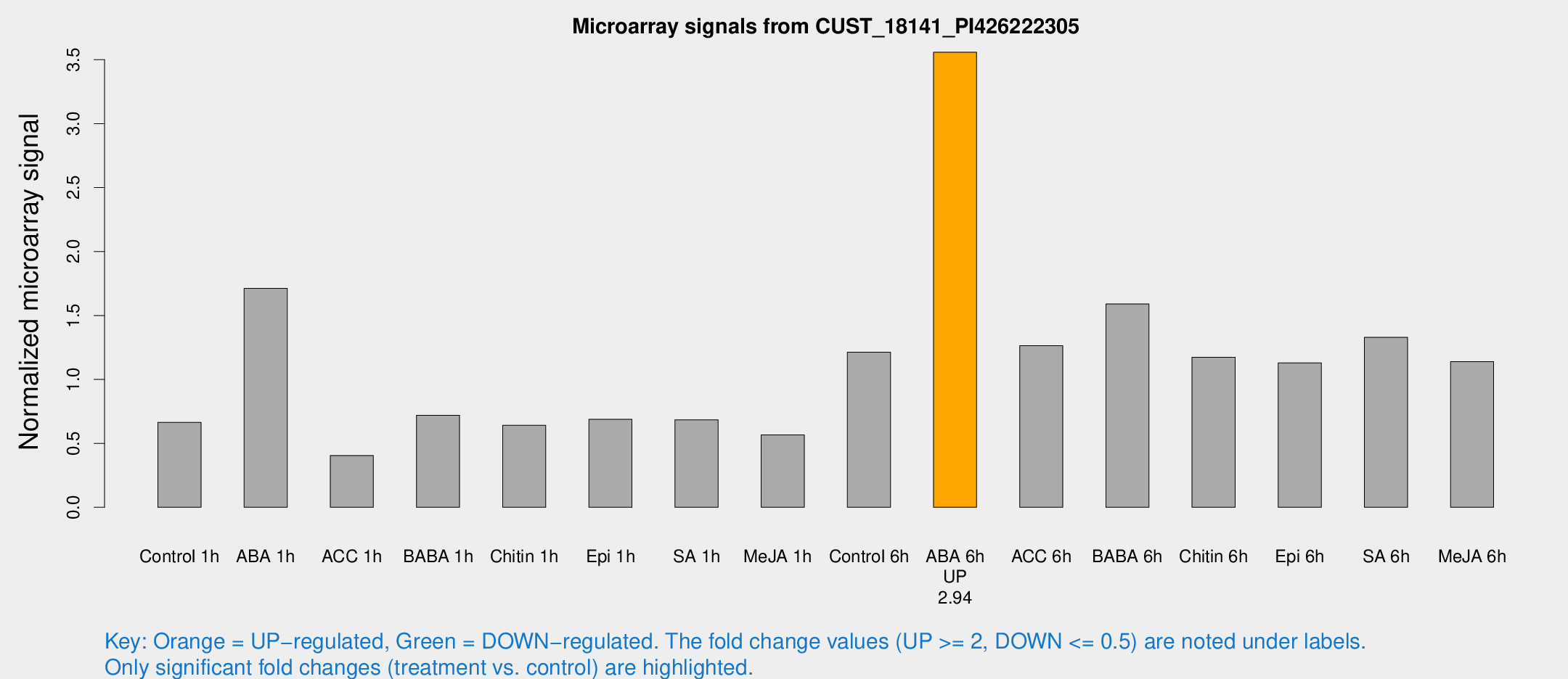
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3458.7 | 250.44 | 0.664289 | 0.0465378 |
| ABA 1h | 8459.74 | 2038.05 | 1.71263 | 0.421123 |
| ACC 1h | 2337.51 | 596.982 | 0.404768 | 0.095785 |
| BABA 1h | 3958.12 | 1065.91 | 0.720074 | 0.16852 |
| Chitin 1h | 3065.92 | 487.437 | 0.641979 | 0.0857 |
| Epi 1h | 3190.02 | 553.642 | 0.687752 | 0.119741 |
| SA 1h | 3795.36 | 815.993 | 0.683473 | 0.112686 |
| Me-JA 1h | 2475.94 | 478.119 | 0.565608 | 0.0738192 |
| Control 6h | 6641.09 | 1553.1 | 1.21233 | 0.217907 |
| ABA 6h | 19985.3 | 2734.4 | 3.55833 | 0.318553 |
| ACC 6h | 7725.53 | 1273.58 | 1.26368 | 0.10061 |
| BABA 6h | 9357.48 | 1138.82 | 1.58942 | 0.157671 |
| Chitin 6h | 6630.71 | 1073.87 | 1.17377 | 0.150773 |
| Epi 6h | 6685.09 | 740.49 | 1.12945 | 0.0653598 |
| SA 6h | 7354.07 | 1868.38 | 1.32847 | 0.239455 |
| Me-JA 6h | 6630.77 | 1936.66 | 1.13874 | 0.364704 |
Source Transcript PGSC0003DMT400048452 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G08570.1 | +1 | 6e-86 | 269 | 147/209 (70%) | atypical CYS HIS rich thioredoxin 4 | chr1:2713059-2714312 FORWARD LENGTH=275 |
