Probe CUST_17552_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17552_PI426222305 | JHI_St_60k_v1 | DMT400008321 | ATTGATCAACAGATGAACGGTGTTGATTTGAGAAGAAATGGAGGGGTTGTTGATGAGGAT |
All Microarray Probes Designed to Gene DMG401003209
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17552_PI426222305 | JHI_St_60k_v1 | DMT400008321 | ATTGATCAACAGATGAACGGTGTTGATTTGAGAAGAAATGGAGGGGTTGTTGATGAGGAT |
| CUST_17476_PI426222305 | JHI_St_60k_v1 | DMT400008327 | GCCCTGTTTTTGCTCTTCTTTAATTCCTCCCAGCTCACGATCTTTGAGTTTGGGTTTTGT |
Microarray Signals from CUST_17552_PI426222305
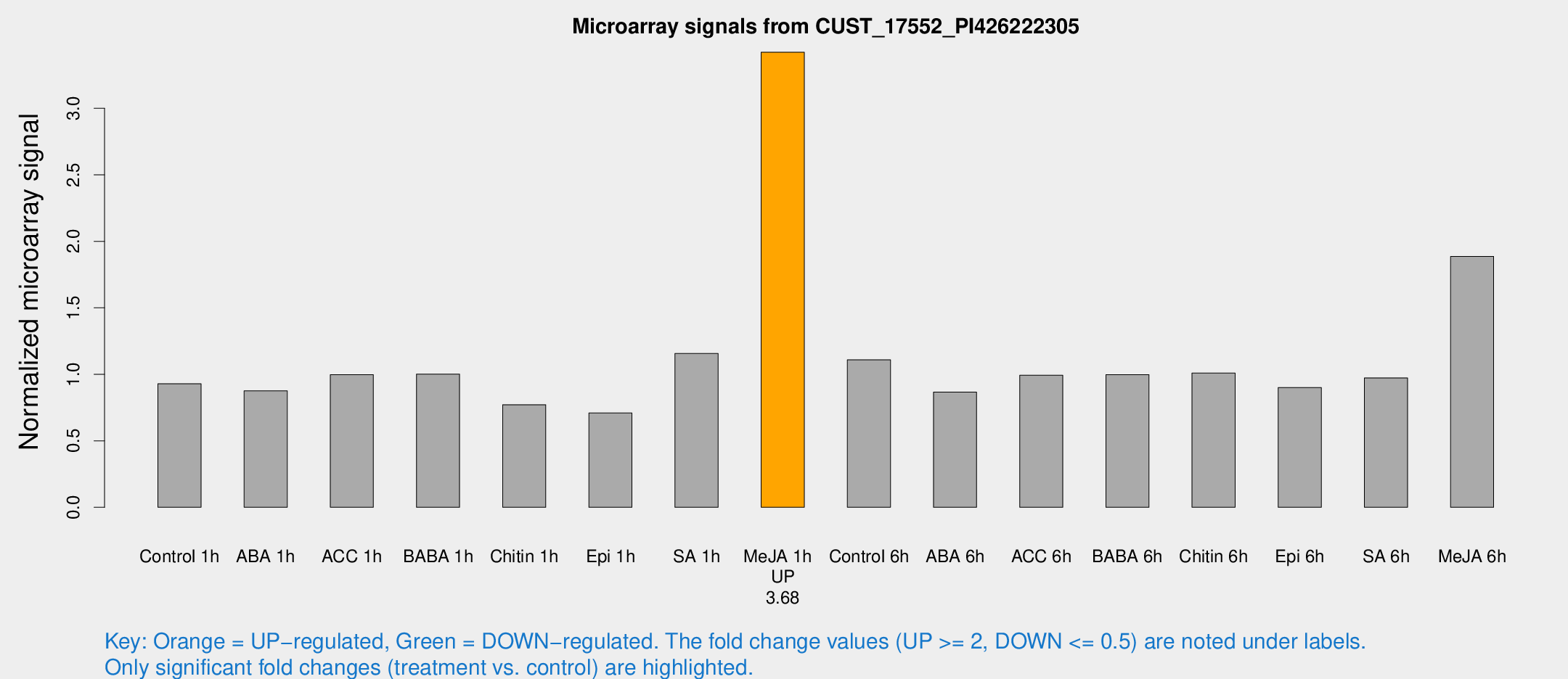
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 141.462 | 15.8043 | 0.928768 | 0.0772394 |
| ABA 1h | 121.983 | 26.8907 | 0.875476 | 0.126354 |
| ACC 1h | 165.895 | 46.8853 | 0.996899 | 0.236135 |
| BABA 1h | 147.414 | 17.4858 | 1.00132 | 0.0638335 |
| Chitin 1h | 104.484 | 6.96236 | 0.771272 | 0.0512853 |
| Epi 1h | 94.5505 | 13.7464 | 0.709032 | 0.105522 |
| SA 1h | 208.308 | 82.5885 | 1.15663 | 0.539384 |
| Me-JA 1h | 421.847 | 25.5188 | 3.42149 | 0.199643 |
| Control 6h | 171.4 | 25.2407 | 1.10898 | 0.080481 |
| ABA 6h | 142.75 | 23.0034 | 0.865995 | 0.122151 |
| ACC 6h | 177.346 | 33.9934 | 0.993246 | 0.0625831 |
| BABA 6h | 168.875 | 11.5014 | 0.997689 | 0.0627481 |
| Chitin 6h | 162.261 | 10.4495 | 1.00879 | 0.0639825 |
| Epi 6h | 161.897 | 38.6332 | 0.900594 | 0.266747 |
| SA 6h | 151.658 | 29.711 | 0.972794 | 0.103264 |
| Me-JA 6h | 298.975 | 63.5839 | 1.88636 | 0.33338 |
Source Transcript PGSC0003DMT400008321 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G10610.1 | +1 | 1e-131 | 381 | 197/268 (74%) | CTC-interacting domain 12 | chr4:6557336-6559143 FORWARD LENGTH=336 |
