Probe CUST_17276_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17276_PI426222305 | JHI_St_60k_v1 | DMT400057934 | CTTAAGAGGATGGTTCTCAGTTCATCAGAATTGTTCAGCTCTGGTTGTATCTCTTCCGAT |
All Microarray Probes Designed to Gene DMG400022500
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17276_PI426222305 | JHI_St_60k_v1 | DMT400057934 | CTTAAGAGGATGGTTCTCAGTTCATCAGAATTGTTCAGCTCTGGTTGTATCTCTTCCGAT |
Microarray Signals from CUST_17276_PI426222305
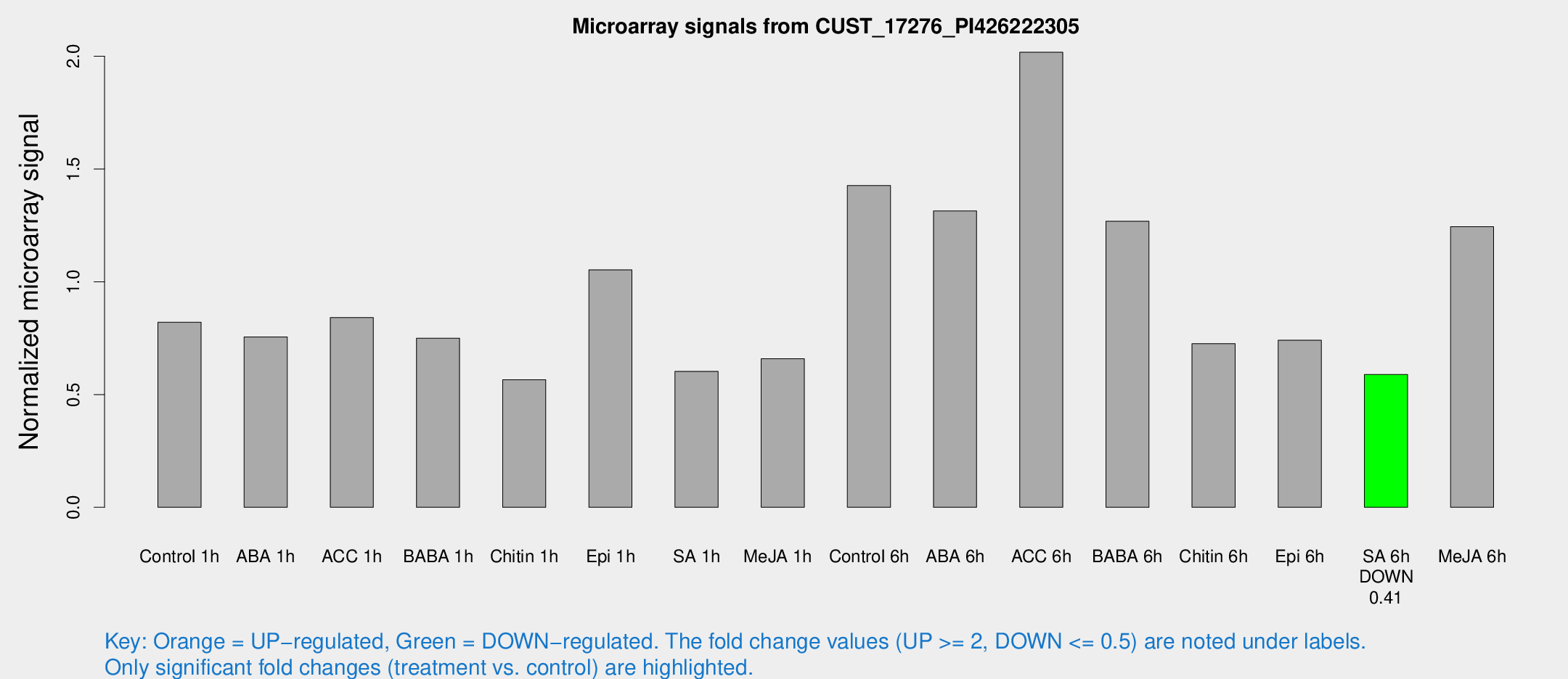
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.1303 | 4.30537 | 0.820511 | 0.292964 |
| ABA 1h | 10.2823 | 3.31029 | 0.754985 | 0.302051 |
| ACC 1h | 12.7306 | 3.69785 | 0.841175 | 0.278555 |
| BABA 1h | 10.883 | 3.59082 | 0.749902 | 0.290798 |
| Chitin 1h | 7.28073 | 3.41718 | 0.565867 | 0.273119 |
| Epi 1h | 13.6667 | 3.40688 | 1.05257 | 0.306712 |
| SA 1h | 10.5279 | 4.89433 | 0.602615 | 0.278895 |
| Me-JA 1h | 7.89668 | 3.48098 | 0.658698 | 0.322101 |
| Control 6h | 21.1948 | 5.20462 | 1.42656 | 0.300136 |
| ABA 6h | 20.0207 | 3.8505 | 1.31415 | 0.267479 |
| ACC 6h | 36.1248 | 12.6734 | 2.01782 | 0.386247 |
| BABA 6h | 20.015 | 4.17695 | 1.26811 | 0.266724 |
| Chitin 6h | 11.6911 | 3.9581 | 0.725843 | 0.294316 |
| Epi 6h | 15.4377 | 8.34608 | 0.740629 | 0.448983 |
| SA 6h | 8.23365 | 3.76496 | 0.589096 | 0.277845 |
| Me-JA 6h | 19.8045 | 7.27455 | 1.24405 | 0.455009 |
Source Transcript PGSC0003DMT400057934 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G30230.2 | +1 | 3e-85 | 261 | 187/230 (81%) | Glutathione S-transferase, C-terminal-like;Translation elongation factor EF1B/ribosomal protein S6 | chr1:10639286-10640601 FORWARD LENGTH=260 |
