Probe CUST_16593_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16593_PI426222305 | JHI_St_60k_v1 | DMT400069547 | CACGATTTACGCTAGTTTGTTATTCGAATTTGAGCAGGTCATTATGTATCAACTATGGTA |
All Microarray Probes Designed to Gene DMG400027035
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16659_PI426222305 | JHI_St_60k_v1 | DMT400069546 | CACGATTTACGCTAGTTTGTTATTCGAATTTGAGCAGGTCATTATGTATCAACTATGGTA |
| CUST_16593_PI426222305 | JHI_St_60k_v1 | DMT400069547 | CACGATTTACGCTAGTTTGTTATTCGAATTTGAGCAGGTCATTATGTATCAACTATGGTA |
Microarray Signals from CUST_16593_PI426222305
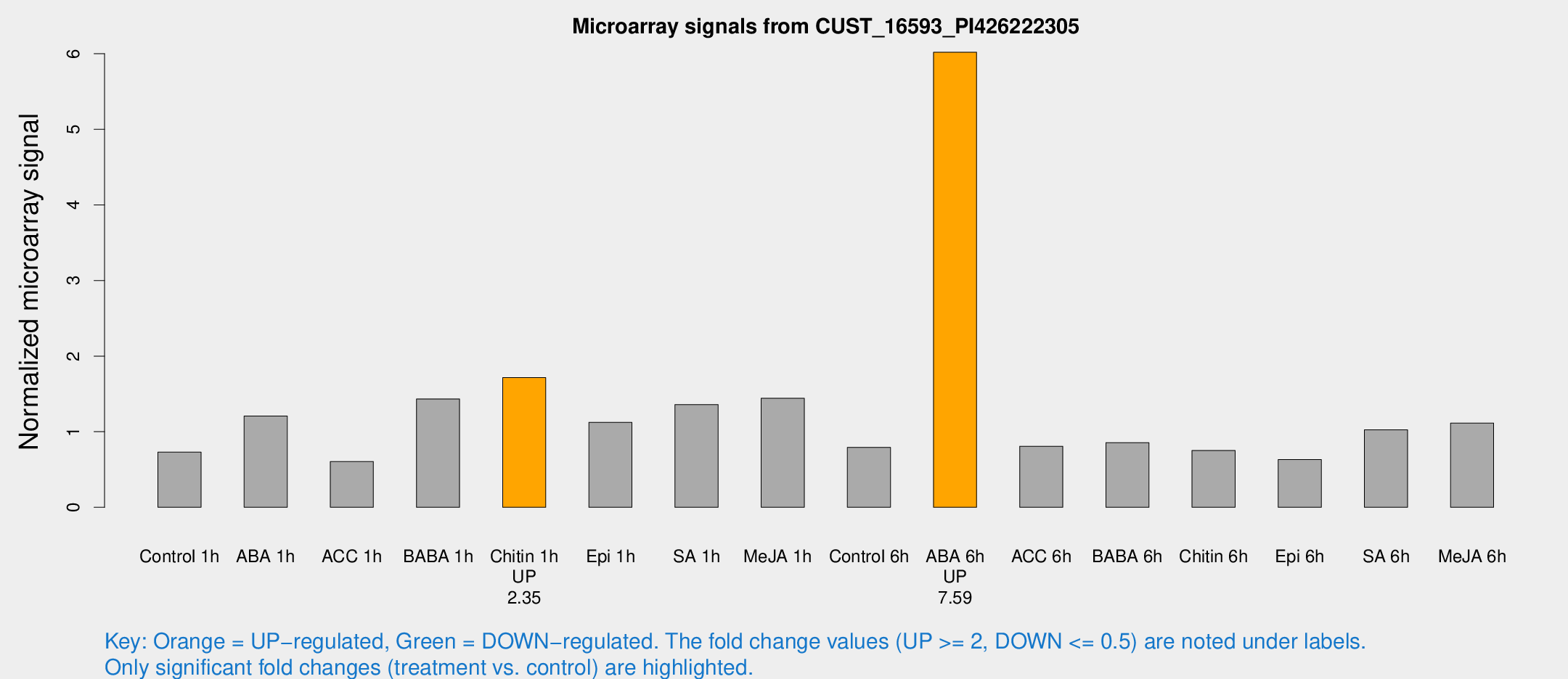
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 231.386 | 18.7156 | 0.728943 | 0.0434093 |
| ABA 1h | 357.482 | 80.5745 | 1.20723 | 0.265855 |
| ACC 1h | 219.171 | 65.3737 | 0.605238 | 0.179985 |
| BABA 1h | 450.579 | 76.7936 | 1.4329 | 0.122808 |
| Chitin 1h | 515.388 | 122.594 | 1.71565 | 0.371325 |
| Epi 1h | 328.196 | 76.0917 | 1.12382 | 0.307838 |
| SA 1h | 458.673 | 93.601 | 1.35771 | 0.250161 |
| Me-JA 1h | 375.588 | 38.172 | 1.44243 | 0.0843275 |
| Control 6h | 260.566 | 48.6589 | 0.792608 | 0.0980447 |
| ABA 6h | 2107.83 | 420.819 | 6.01907 | 0.969361 |
| ACC 6h | 299.524 | 44.7739 | 0.8071 | 0.0931699 |
| BABA 6h | 306.732 | 36.7571 | 0.854985 | 0.0783257 |
| Chitin 6h | 253.247 | 16.3322 | 0.751064 | 0.0448515 |
| Epi 6h | 233.776 | 44.1666 | 0.631508 | 0.082832 |
| SA 6h | 322.847 | 28.9014 | 1.02586 | 0.060423 |
| Me-JA 6h | 353.218 | 33.3838 | 1.11373 | 0.0652685 |
Source Transcript PGSC0003DMT400069547 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G17720.1 | +2 | 3e-95 | 294 | 166/271 (61%) | RNA-binding (RRM/RBD/RNP motifs) family protein | chr4:9862660-9864498 REVERSE LENGTH=313 |
