Probe CUST_15643_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15643_PI426222305 | JHI_St_60k_v1 | DMT400011286 | TCAAGTCGGAGCTAGAAGTTTGGGTATGGATTTGATCGAACCAGTAACTTTTGATCAAAT |
All Microarray Probes Designed to Gene DMG400004418
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15643_PI426222305 | JHI_St_60k_v1 | DMT400011286 | TCAAGTCGGAGCTAGAAGTTTGGGTATGGATTTGATCGAACCAGTAACTTTTGATCAAAT |
| CUST_15613_PI426222305 | JHI_St_60k_v1 | DMT400011285 | TCAGACCATTACCATCATCTCATCATCAAGGTTTTCAAGCAAATCTGGTTTGATATTCAA |
Microarray Signals from CUST_15643_PI426222305
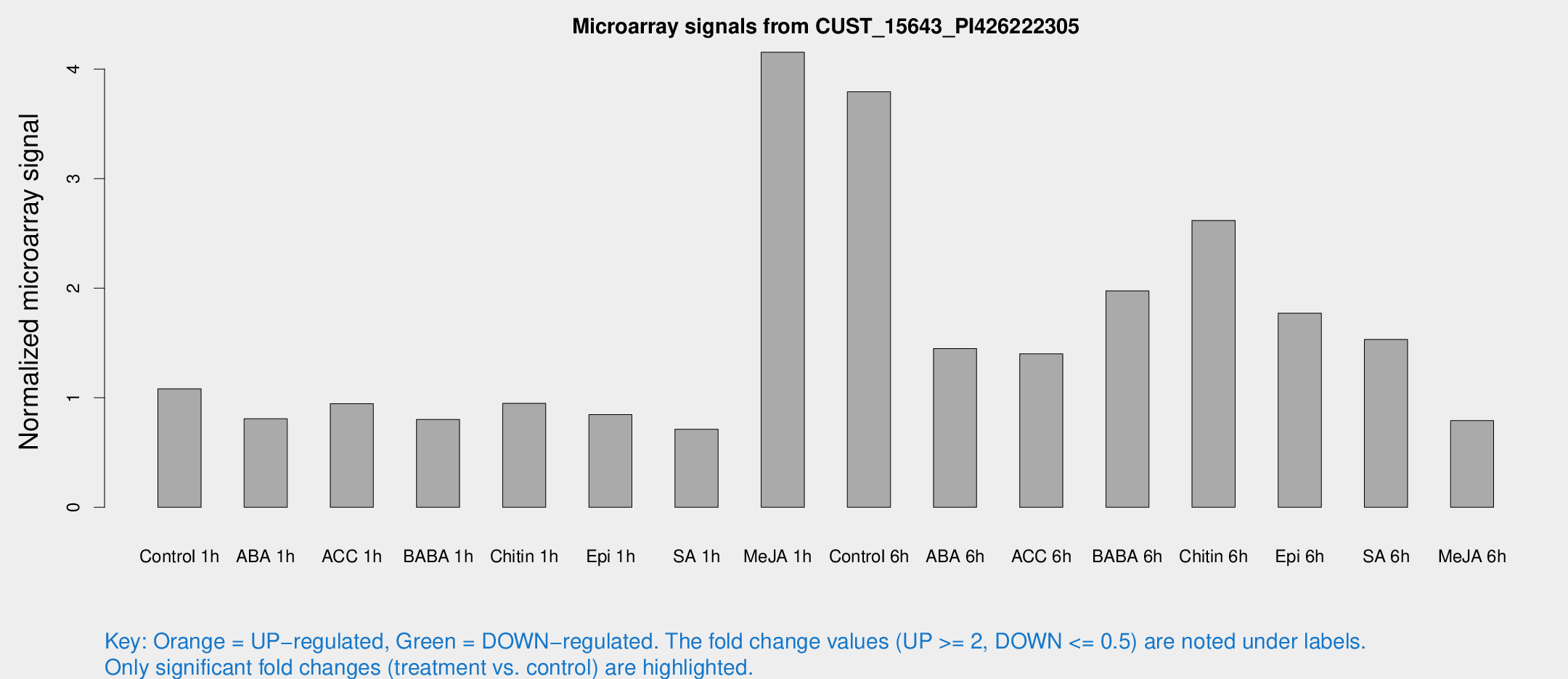
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.28943 | 3.16253 | 1.08089 | 0.453531 |
| ABA 1h | 5.28708 | 3.06432 | 0.80829 | 0.468167 |
| ACC 1h | 7.43669 | 3.45982 | 0.945706 | 0.473293 |
| BABA 1h | 5.725 | 3.3269 | 0.80174 | 0.464767 |
| Chitin 1h | 6.34993 | 3.22681 | 0.948839 | 0.486852 |
| Epi 1h | 5.37017 | 3.1172 | 0.845715 | 0.485045 |
| SA 1h | 5.41157 | 3.13789 | 0.712025 | 0.412364 |
| Me-JA 1h | 25.5971 | 3.59776 | 4.15334 | 0.818061 |
| Control 6h | 51.1103 | 33.107 | 3.79311 | 4.93181 |
| ABA 6h | 13.1959 | 5.15066 | 1.44919 | 0.727547 |
| ACC 6h | 13.9691 | 5.90199 | 1.401 | 0.498721 |
| BABA 6h | 17.7222 | 4.52482 | 1.97513 | 0.554304 |
| Chitin 6h | 23.2207 | 8.35846 | 2.61804 | 0.950373 |
| Epi 6h | 16.191 | 4.36965 | 1.77172 | 0.784443 |
| SA 6h | 12.7618 | 4.46518 | 1.53215 | 0.874538 |
| Me-JA 6h | 5.82973 | 3.38585 | 0.791324 | 0.458966 |
Source Transcript PGSC0003DMT400011286 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G06920.1 | +1 | 5e-26 | 106 | 94/308 (31%) | ovate family protein 4 | chr1:2124854-2125801 REVERSE LENGTH=315 |
