Probe CUST_14714_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14714_PI426222305 | JHI_St_60k_v1 | DMT400066556 | CAAGCTTATGGTGGGAAACCAAAGTCTTAGTTGTGTCAGTCAATGTTTCTGAGATTTTTT |
All Microarray Probes Designed to Gene DMG400025867
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14714_PI426222305 | JHI_St_60k_v1 | DMT400066556 | CAAGCTTATGGTGGGAAACCAAAGTCTTAGTTGTGTCAGTCAATGTTTCTGAGATTTTTT |
| CUST_14578_PI426222305 | JHI_St_60k_v1 | DMT400066555 | TTGAAATCACAGAAATCTACGTCGATTTGTCAGCAAGCTTATGGTGGGAAACCAAAGTCT |
Microarray Signals from CUST_14714_PI426222305
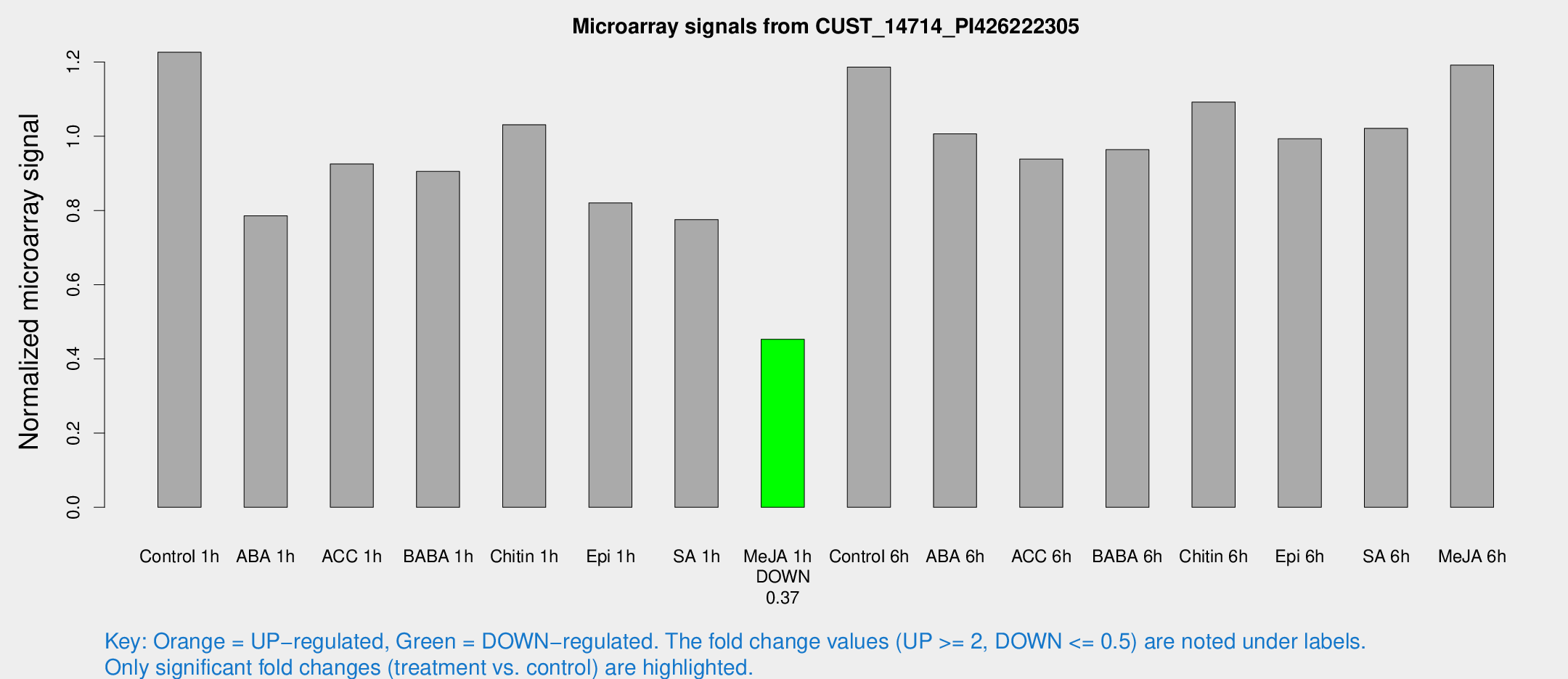
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3312.39 | 445.92 | 1.22653 | 0.102081 |
| ABA 1h | 1847.09 | 111.628 | 0.785614 | 0.0453779 |
| ACC 1h | 2684.23 | 618.51 | 0.925437 | 0.176937 |
| BABA 1h | 2420.27 | 506.185 | 0.905232 | 0.118199 |
| Chitin 1h | 2489 | 280.131 | 1.03108 | 0.0595457 |
| Epi 1h | 1888.48 | 122.555 | 0.820508 | 0.0512935 |
| SA 1h | 2170.62 | 343.759 | 0.775486 | 0.100949 |
| Me-JA 1h | 984.955 | 75.9597 | 0.452796 | 0.0261885 |
| Control 6h | 3271.43 | 617.406 | 1.18642 | 0.15158 |
| ABA 6h | 3141.01 | 860.373 | 1.00649 | 0.383871 |
| ACC 6h | 3075.96 | 769.764 | 0.938727 | 0.232042 |
| BABA 6h | 2877.64 | 237.201 | 0.963879 | 0.0556642 |
| Chitin 6h | 3096.4 | 237.053 | 1.09234 | 0.0997505 |
| Epi 6h | 2979.74 | 198.399 | 0.993551 | 0.0573775 |
| SA 6h | 2836.08 | 615.933 | 1.02135 | 0.139633 |
| Me-JA 6h | 3195.2 | 412.236 | 1.19185 | 0.0688238 |
Source Transcript PGSC0003DMT400066556 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G15320.1 | +2 | 3e-133 | 394 | 218/356 (61%) | Leucine-rich repeat (LRR) family protein | chr2:6666527-6667675 REVERSE LENGTH=382 |
