Probe CUST_14604_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14604_PI426222305 | JHI_St_60k_v1 | DMT400066596 | TTGCTCCAAGTACTTTGACACATGGTATCTAATAAGCCAACACGAGATGATAAGTTTAAC |
All Microarray Probes Designed to Gene DMG400025887
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14689_PI426222305 | JHI_St_60k_v1 | DMT400066595 | TTGCTCCAAGTACTTTGACACATGGTATCTAATAAGCCAACACGAGATGATAAGTTTAAC |
| CUST_14604_PI426222305 | JHI_St_60k_v1 | DMT400066596 | TTGCTCCAAGTACTTTGACACATGGTATCTAATAAGCCAACACGAGATGATAAGTTTAAC |
Microarray Signals from CUST_14604_PI426222305
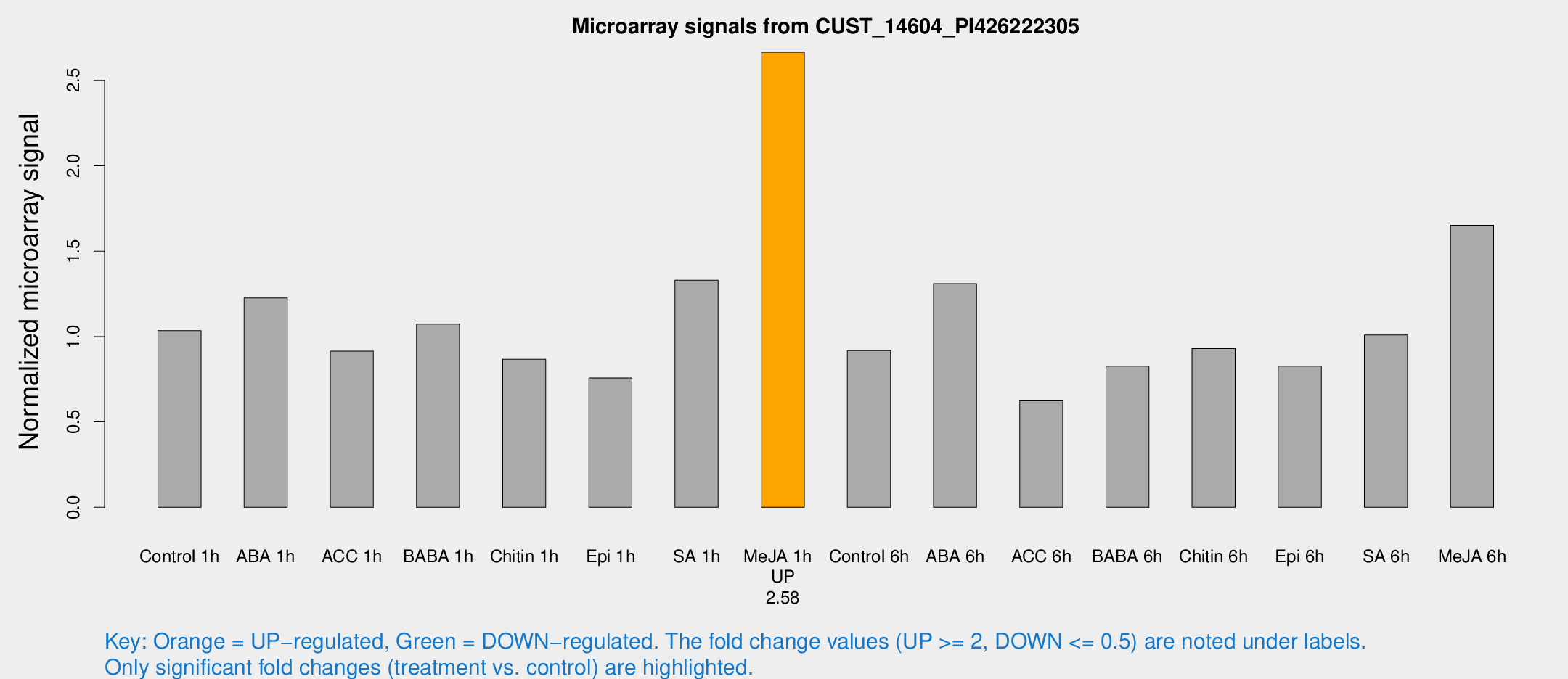
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 103.888 | 16.2844 | 1.03451 | 0.100033 |
| ABA 1h | 119.939 | 38.7051 | 1.22544 | 0.378518 |
| ACC 1h | 100.886 | 31.8697 | 0.91433 | 0.252595 |
| BABA 1h | 105.724 | 19.6101 | 1.07316 | 0.111886 |
| Chitin 1h | 77.3873 | 8.03665 | 0.86649 | 0.0626062 |
| Epi 1h | 67.9052 | 14.4764 | 0.757313 | 0.1818 |
| SA 1h | 136.125 | 17.0495 | 1.32998 | 0.195214 |
| Me-JA 1h | 216.549 | 25.0072 | 2.66488 | 0.159873 |
| Control 6h | 94.1973 | 18.445 | 0.917636 | 0.108717 |
| ABA 6h | 136.763 | 8.63538 | 1.30961 | 0.0826276 |
| ACC 6h | 71.966 | 10.8513 | 0.623921 | 0.135771 |
| BABA 6h | 91.7345 | 9.1938 | 0.826858 | 0.114936 |
| Chitin 6h | 97.5656 | 6.78755 | 0.929499 | 0.0646631 |
| Epi 6h | 92.5262 | 10.1209 | 0.826468 | 0.114208 |
| SA 6h | 101.968 | 19.141 | 1.00921 | 0.0942572 |
| Me-JA 6h | 164.826 | 24.5032 | 1.65166 | 0.129267 |
Source Transcript PGSC0003DMT400066596 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G15570.2 | +1 | 2e-52 | 174 | 83/125 (66%) | Thioredoxin superfamily protein | chr2:6791556-6792902 REVERSE LENGTH=174 |
