Probe CUST_13458_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13458_PI426222305 | JHI_St_60k_v1 | DMT400017633 | AACAAAATGGCAACCACACTGTTCGGAGTCCTATAGTGACATCTACCATTATTGAGGTCT |
All Microarray Probes Designed to Gene DMG400006840
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13578_PI426222305 | JHI_St_60k_v1 | DMT400017634 | CATTATTGAGGTCTGGGGTAGCGAAGACTAAGGGGGAAATAAGAAACACATTATATAAGG |
| CUST_13458_PI426222305 | JHI_St_60k_v1 | DMT400017633 | AACAAAATGGCAACCACACTGTTCGGAGTCCTATAGTGACATCTACCATTATTGAGGTCT |
Microarray Signals from CUST_13458_PI426222305
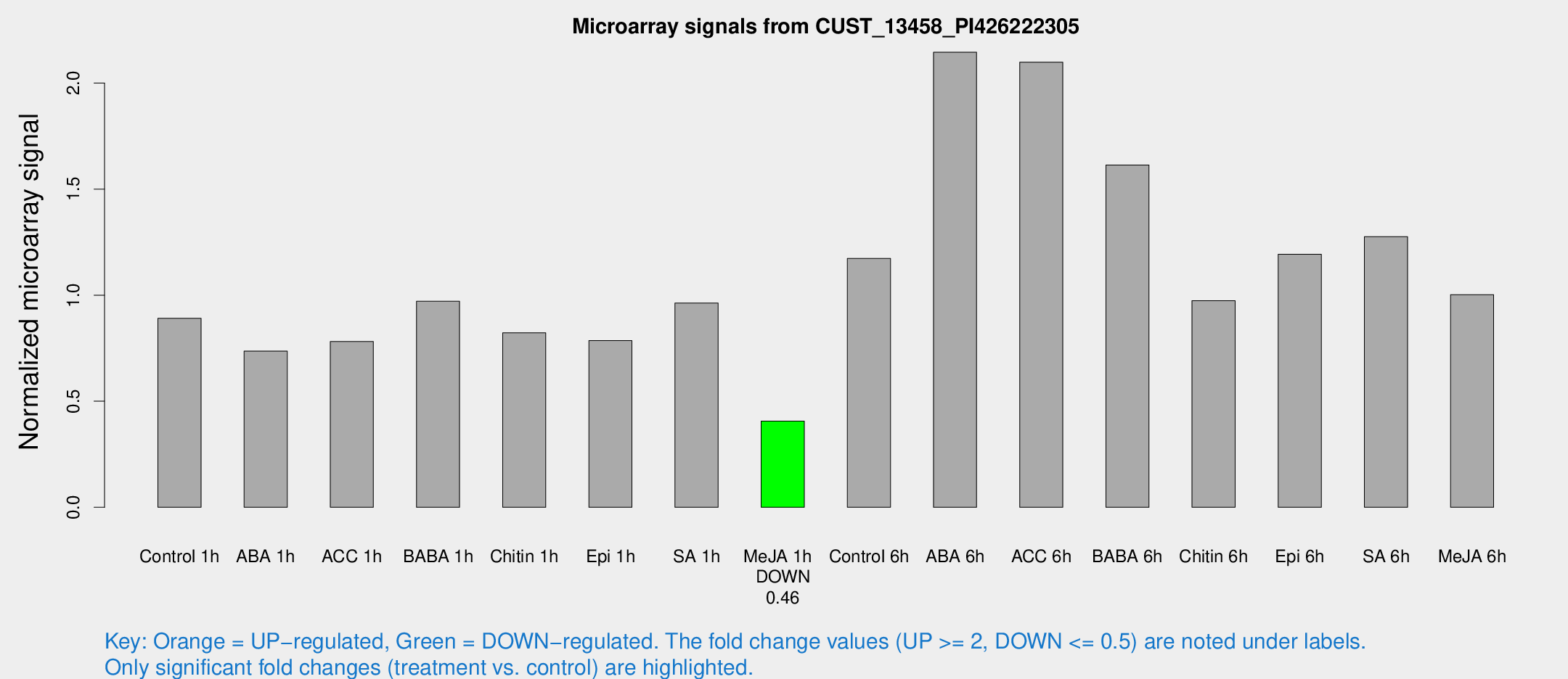
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1317.24 | 214.12 | 0.890783 | 0.101662 |
| ABA 1h | 961.691 | 149.986 | 0.736088 | 0.0614008 |
| ACC 1h | 1220.39 | 255.958 | 0.781442 | 0.131012 |
| BABA 1h | 1476.16 | 392.944 | 0.971121 | 0.212401 |
| Chitin 1h | 1099.86 | 199.989 | 0.822455 | 0.0955044 |
| Epi 1h | 993.846 | 107.41 | 0.786038 | 0.0759534 |
| SA 1h | 1500.12 | 347.19 | 0.962618 | 0.158355 |
| Me-JA 1h | 483.364 | 45.5655 | 0.406543 | 0.023614 |
| Control 6h | 1819.85 | 425.93 | 1.17348 | 0.222118 |
| ABA 6h | 3552.68 | 882.788 | 2.14535 | 0.540143 |
| ACC 6h | 3484.64 | 206.591 | 2.0985 | 0.16868 |
| BABA 6h | 2721.81 | 593.573 | 1.61395 | 0.292173 |
| Chitin 6h | 1502.62 | 98.1663 | 0.974171 | 0.0704478 |
| Epi 6h | 1974.97 | 256.849 | 1.19258 | 0.112955 |
| SA 6h | 1837.85 | 170.76 | 1.27616 | 0.0737159 |
| Me-JA 6h | 1509.94 | 324.606 | 1.00259 | 0.136166 |
Source Transcript PGSC0003DMT400017633 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G34980.1 | +1 | 2e-59 | 204 | 120/289 (42%) | subtilisin-like serine protease 2 | chr4:16656929-16659223 REVERSE LENGTH=764 |
