Probe CUST_13191_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13191_PI426222305 | JHI_St_60k_v1 | DMT400007496 | ATGGTTTTGCTGGAAAATTACAGTGCCCTCAACTATACTGGGCTCGTGAAGATACTCAAG |
All Microarray Probes Designed to Gene DMG400002890
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13191_PI426222305 | JHI_St_60k_v1 | DMT400007496 | ATGGTTTTGCTGGAAAATTACAGTGCCCTCAACTATACTGGGCTCGTGAAGATACTCAAG |
| CUST_13148_PI426222305 | JHI_St_60k_v1 | DMT400007495 | GTGATGAGCCATTCTGATCTATTCCTGTTTTTGCTTATAGGGCTCGTGAAGATACTCAAG |
Microarray Signals from CUST_13191_PI426222305
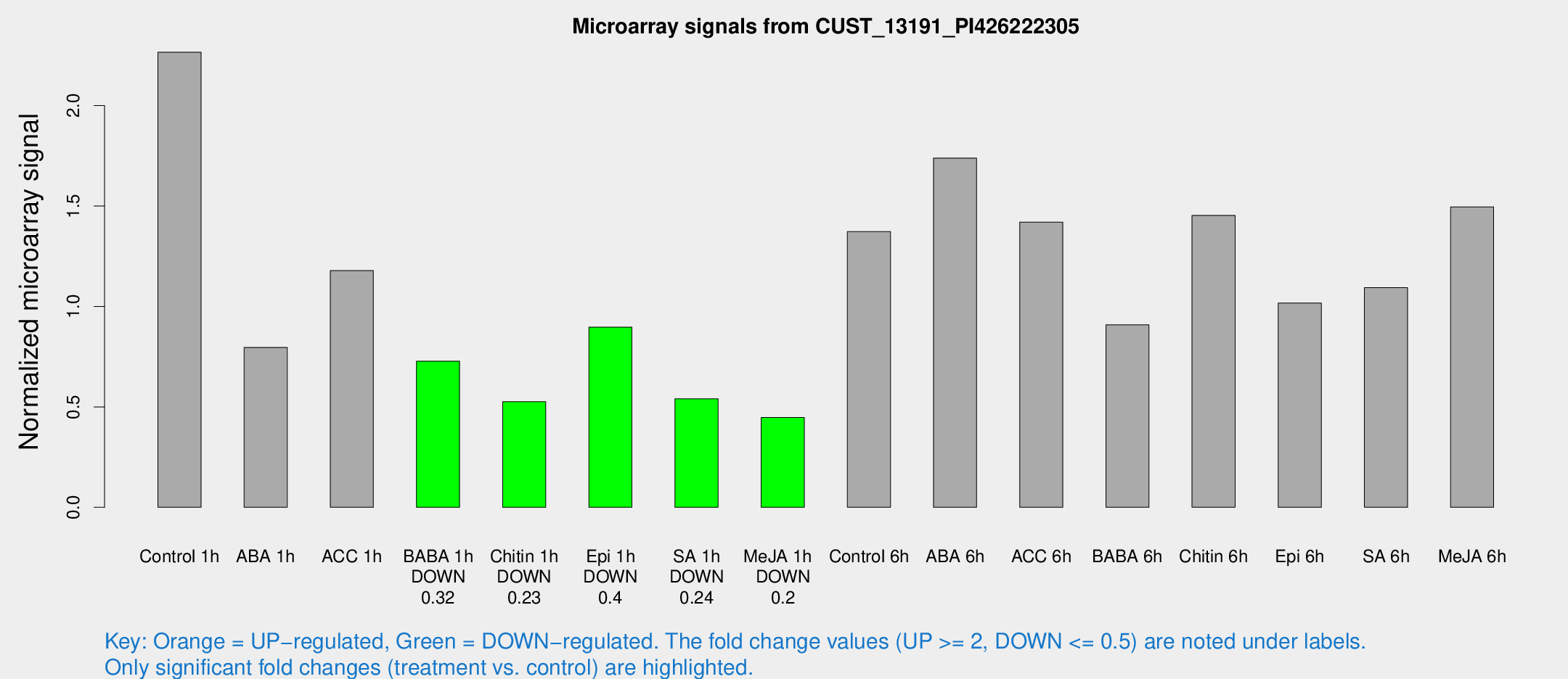
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1133.76 | 226.653 | 2.26585 | 0.373553 |
| ABA 1h | 372.472 | 115.347 | 0.795946 | 0.254021 |
| ACC 1h | 701.181 | 265.109 | 1.17858 | 0.524869 |
| BABA 1h | 372.264 | 106.949 | 0.727232 | 0.184801 |
| Chitin 1h | 232.686 | 41.822 | 0.525394 | 0.0695084 |
| Epi 1h | 376.471 | 44.574 | 0.896735 | 0.0986591 |
| SA 1h | 280.933 | 68.6815 | 0.540066 | 0.0987248 |
| Me-JA 1h | 176.306 | 18.9922 | 0.446981 | 0.0271593 |
| Control 6h | 808.768 | 378.411 | 1.37271 | 0.616743 |
| ABA 6h | 976.963 | 269.048 | 1.73858 | 0.539669 |
| ACC 6h | 818.237 | 191.942 | 1.41974 | 0.116595 |
| BABA 6h | 516.488 | 128.473 | 0.908331 | 0.212668 |
| Chitin 6h | 750.158 | 87.6518 | 1.45349 | 0.194022 |
| Epi 6h | 583.164 | 131.692 | 1.01708 | 0.197435 |
| SA 6h | 575.895 | 192.124 | 1.09379 | 0.276185 |
| Me-JA 6h | 750.442 | 168.44 | 1.49597 | 0.22556 |
Source Transcript PGSC0003DMT400007496 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G26660.1 | +3 | 2e-107 | 322 | 199/298 (67%) | SPX domain gene 2 | chr2:11338932-11340703 FORWARD LENGTH=287 |
