Probe CUST_13188_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13188_PI426222305 | JHI_St_60k_v1 | DMT400007516 | TAACACCGCAAGAGAGTCAAAAGTTCGTGTCAAGTCAAAACTAGACAAACAAATTCAAAC |
All Microarray Probes Designed to Gene DMG400002901
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13188_PI426222305 | JHI_St_60k_v1 | DMT400007516 | TAACACCGCAAGAGAGTCAAAAGTTCGTGTCAAGTCAAAACTAGACAAACAAATTCAAAC |
| CUST_13080_PI426222305 | JHI_St_60k_v1 | DMT400007514 | ACCGCAAGAGAGTCAAAAGTTCGTGTCAAGTCAAAACTAGACAAACAAATTCAAACGAAG |
Microarray Signals from CUST_13188_PI426222305
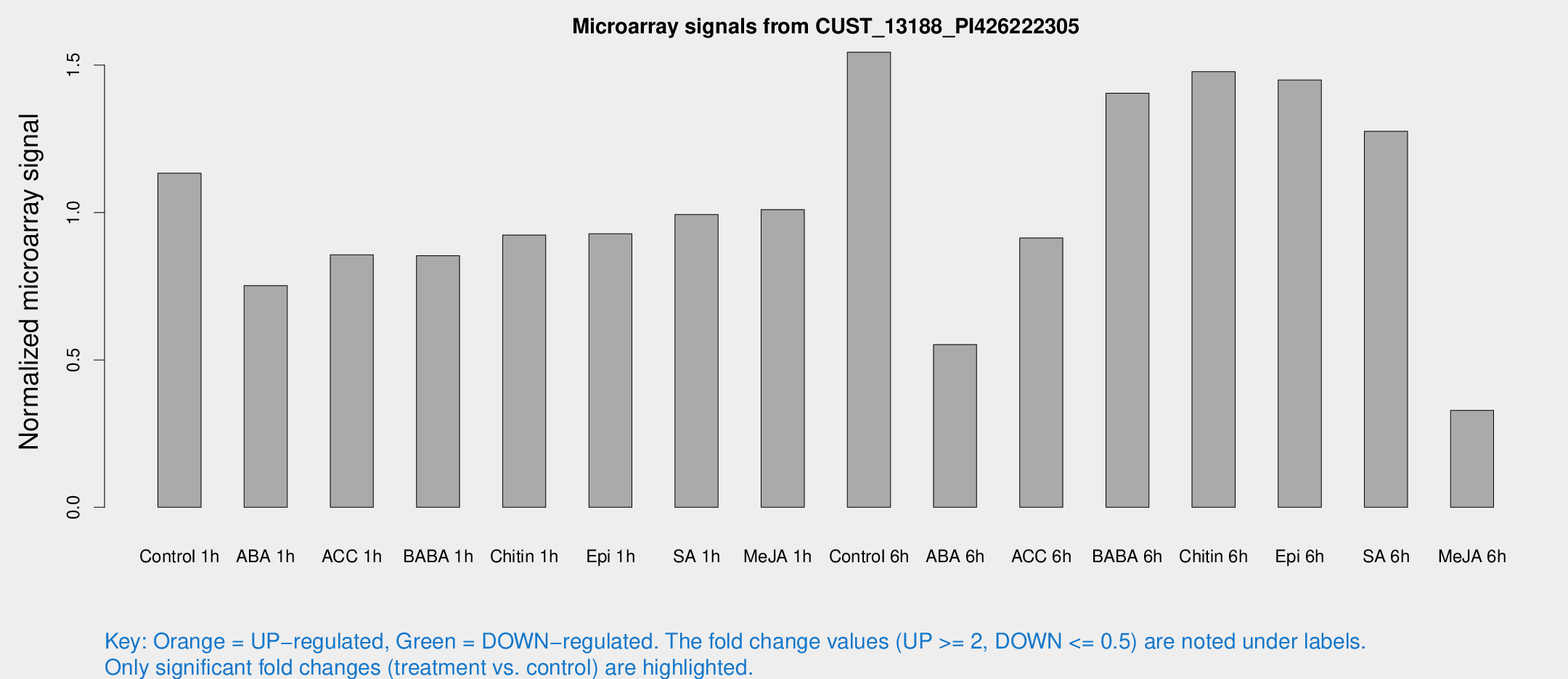
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 73.2825 | 7.56614 | 1.13355 | 0.0853282 |
| ABA 1h | 44.6401 | 10.2191 | 0.752301 | 0.121395 |
| ACC 1h | 56.4499 | 4.88876 | 0.856342 | 0.0747491 |
| BABA 1h | 54.4594 | 9.68052 | 0.853424 | 0.0827869 |
| Chitin 1h | 54.6632 | 9.67854 | 0.923592 | 0.177891 |
| Epi 1h | 51.5656 | 4.46676 | 0.927774 | 0.0810673 |
| SA 1h | 65.427 | 5.07159 | 0.993314 | 0.0769313 |
| Me-JA 1h | 53.0117 | 4.71823 | 1.01011 | 0.0899782 |
| Control 6h | 102.985 | 19.6062 | 1.54387 | 0.198425 |
| ABA 6h | 40.1291 | 10.8738 | 0.552087 | 0.174766 |
| ACC 6h | 71.3721 | 18.5243 | 0.913843 | 0.100294 |
| BABA 6h | 104.707 | 20.2502 | 1.40494 | 0.329114 |
| Chitin 6h | 102.543 | 14.442 | 1.47798 | 0.156916 |
| Epi 6h | 105.504 | 10.4813 | 1.44947 | 0.146552 |
| SA 6h | 81.5308 | 8.04395 | 1.27547 | 0.0946962 |
| Me-JA 6h | 25.5726 | 11.4702 | 0.329029 | 0.131385 |
Source Transcript PGSC0003DMT400007516 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G19150.1 | +3 | 2e-106 | 318 | 146/176 (83%) | photosystem I light harvesting complex gene 6 | chr1:6612806-6613799 FORWARD LENGTH=270 |
