Probe CUST_12617_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12617_PI426222305 | JHI_St_60k_v1 | DMT400063718 | TTTGACCAAAAGGGATTCACTGGTGGACTAAACTACTACCGCGCTCTTGATTTGAACTGG |
All Microarray Probes Designed to Gene DMG400024765
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12617_PI426222305 | JHI_St_60k_v1 | DMT400063718 | TTTGACCAAAAGGGATTCACTGGTGGACTAAACTACTACCGCGCTCTTGATTTGAACTGG |
| CUST_12320_PI426222305 | JHI_St_60k_v1 | DMT400063719 | TTTATATGTTTCTTATACCGTACCGTTGAATTTTGGGCAGGAACTGGGAGCTGACAGCCC |
Microarray Signals from CUST_12617_PI426222305
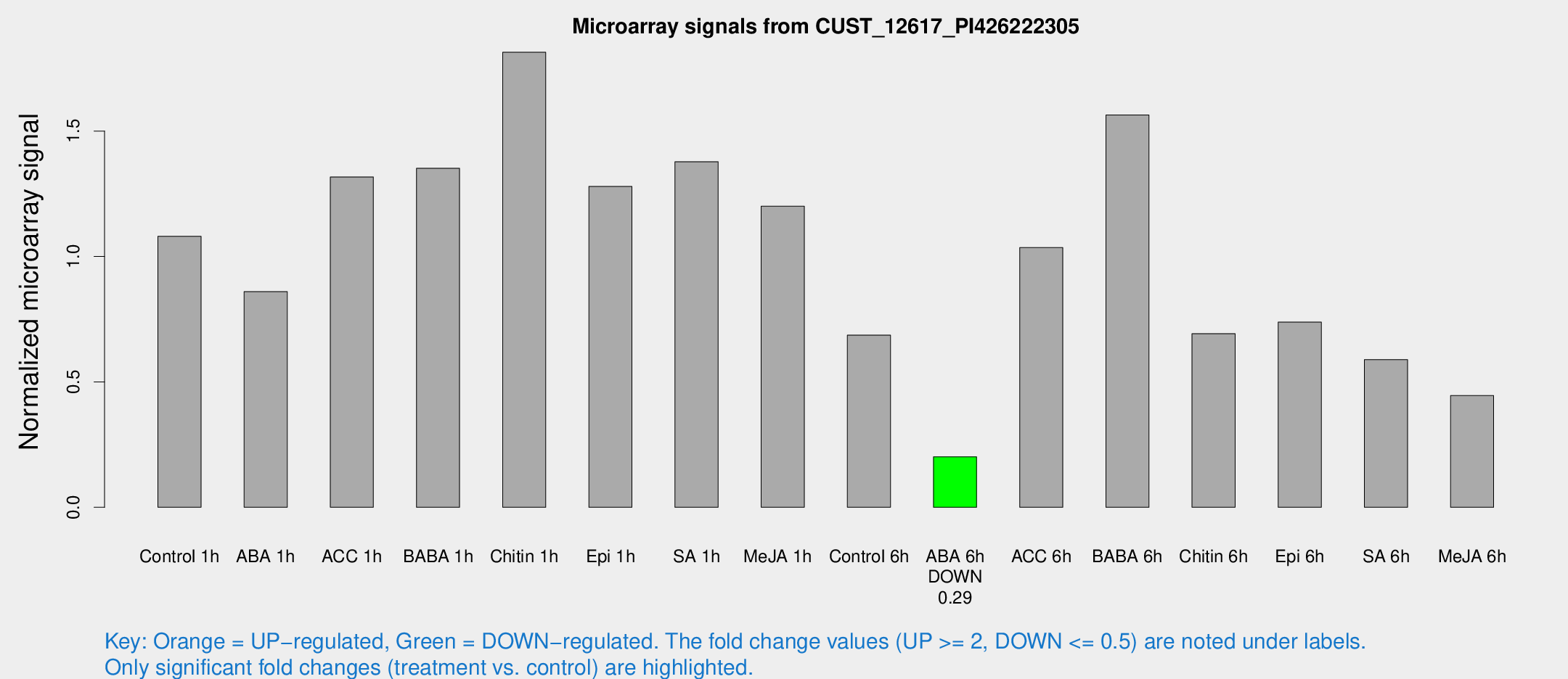
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 808.918 | 120.455 | 1.08072 | 0.116701 |
| ABA 1h | 592.968 | 159.931 | 0.85992 | 0.167387 |
| ACC 1h | 1017.15 | 181.939 | 1.31714 | 0.20134 |
| BABA 1h | 1015.98 | 241.434 | 1.35178 | 0.229542 |
| Chitin 1h | 1208.27 | 146.529 | 1.81437 | 0.207086 |
| Epi 1h | 820.239 | 94.9469 | 1.27931 | 0.152172 |
| SA 1h | 1059.47 | 174.382 | 1.37797 | 0.144959 |
| Me-JA 1h | 720.681 | 60.9762 | 1.20044 | 0.0695654 |
| Control 6h | 575.796 | 176.354 | 0.686606 | 0.225787 |
| ABA 6h | 157.524 | 12.8348 | 0.201564 | 0.0188006 |
| ACC 6h | 876.099 | 87.4258 | 1.0356 | 0.101612 |
| BABA 6h | 1289.19 | 122.111 | 1.5639 | 0.134568 |
| Chitin 6h | 544.118 | 58.1695 | 0.692423 | 0.0685326 |
| Epi 6h | 620.339 | 92.6449 | 0.738204 | 0.121813 |
| SA 6h | 497.59 | 160.998 | 0.588823 | 0.200508 |
| Me-JA 6h | 348.864 | 91.5677 | 0.445432 | 0.088524 |
Source Transcript PGSC0003DMT400063718 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G05600.1 | +3 | 7e-143 | 413 | 201/331 (61%) | alpha/beta-Hydrolases superfamily protein | chr3:1623485-1624704 REVERSE LENGTH=331 |
