Probe CUST_12305_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12305_PI426222305 | JHI_St_60k_v1 | DMT400003001 | ACAAGTCCCGTCTTTCACTTTGGAGCCTTCCAAAAGGAATACCCAAGTTGGTTAGATGCA |
All Microarray Probes Designed to Gene DMG400001182
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12474_PI426222305 | JHI_St_60k_v1 | DMT400003003 | AAAGTTGGTTTTTCCATTCCATGTTGGCTGAGAAAATGAGTTAAAATGGTGATTAGCGTG |
| CUST_12305_PI426222305 | JHI_St_60k_v1 | DMT400003001 | ACAAGTCCCGTCTTTCACTTTGGAGCCTTCCAAAAGGAATACCCAAGTTGGTTAGATGCA |
Microarray Signals from CUST_12305_PI426222305
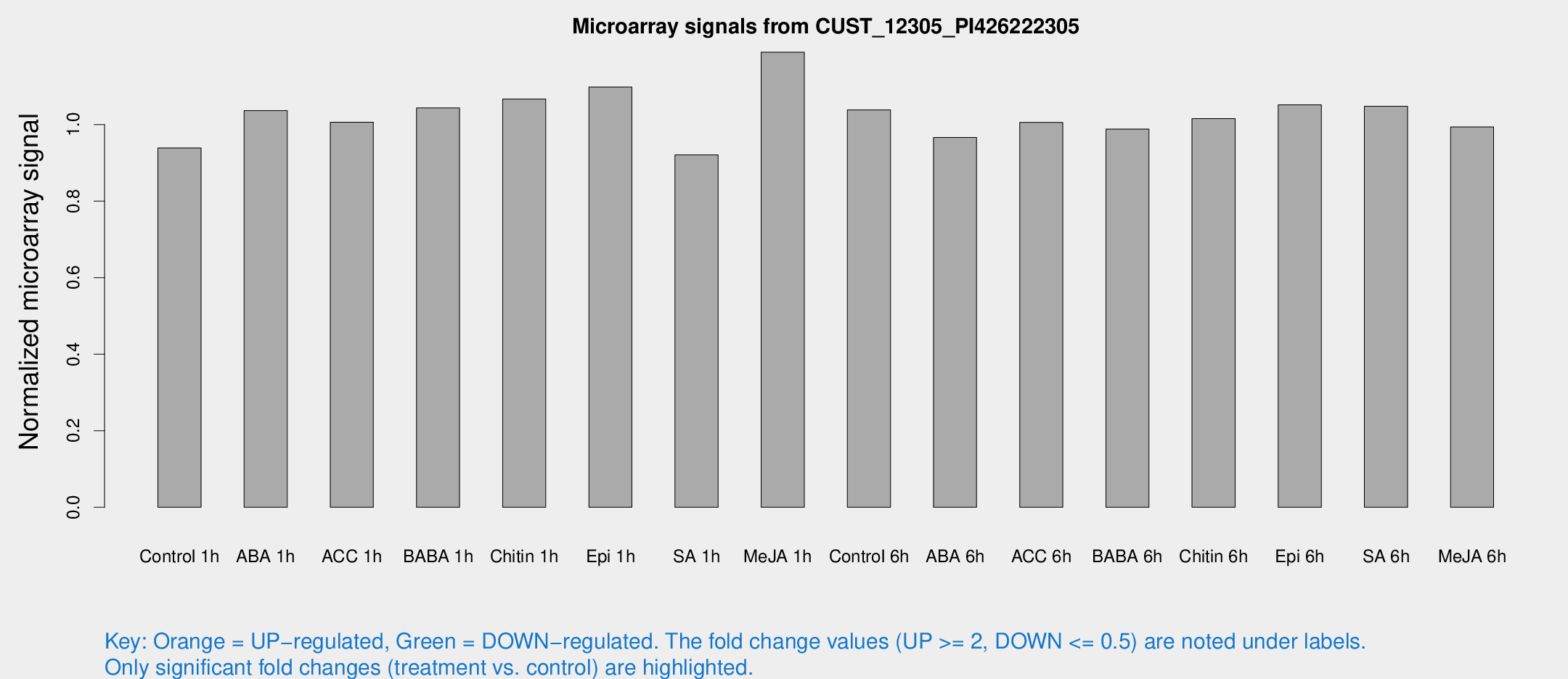
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.79125 | 3.35609 | 0.938951 | 0.543788 |
| ABA 1h | 5.66441 | 3.28787 | 1.03633 | 0.60013 |
| ACC 1h | 6.3864 | 3.70758 | 1.00627 | 0.582705 |
| BABA 1h | 6.20125 | 3.59171 | 1.0436 | 0.604384 |
| Chitin 1h | 5.93222 | 3.44529 | 1.067 | 0.617888 |
| Epi 1h | 5.86841 | 3.40432 | 1.09802 | 0.635832 |
| SA 1h | 5.83447 | 3.38221 | 0.920807 | 0.533423 |
| Me-JA 1h | 5.9939 | 3.48106 | 1.18911 | 0.689584 |
| Control 6h | 6.42892 | 3.56738 | 1.0386 | 0.578242 |
| ABA 6h | 6.34219 | 3.67694 | 0.966588 | 0.559712 |
| ACC 6h | 7.31749 | 4.36577 | 1.00578 | 0.582415 |
| BABA 6h | 6.83742 | 3.97194 | 0.988253 | 0.572501 |
| Chitin 6h | 6.66516 | 3.86344 | 1.01545 | 0.588386 |
| Epi 6h | 7.43306 | 4.11691 | 1.05158 | 0.584375 |
| SA 6h | 6.39497 | 3.70571 | 1.04812 | 0.606948 |
| Me-JA 6h | 6.10552 | 3.5384 | 0.9938 | 0.575469 |
Source Transcript PGSC0003DMT400003001 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23780.1 | +1 | 1e-63 | 202 | 102/169 (60%) | RING/U-box superfamily protein | chr2:10123551-10124234 REVERSE LENGTH=227 |
