Probe CUST_11127_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11127_PI426222305 | JHI_St_60k_v1 | DMT400078452 | CCGCACACACTTGTGAGCATTTTTGAAAATTTTCTCTATGGATTATCCTGGCTTTAAATA |
All Microarray Probes Designed to Gene DMG400030536
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11052_PI426222305 | JHI_St_60k_v1 | DMT400078451 | CCGGAGGAAGGTGTCAAACAGATAATGCTACTAATAGATTTCTTTGTTCTGATACCAATG |
| CUST_11127_PI426222305 | JHI_St_60k_v1 | DMT400078452 | CCGCACACACTTGTGAGCATTTTTGAAAATTTTCTCTATGGATTATCCTGGCTTTAAATA |
Microarray Signals from CUST_11127_PI426222305
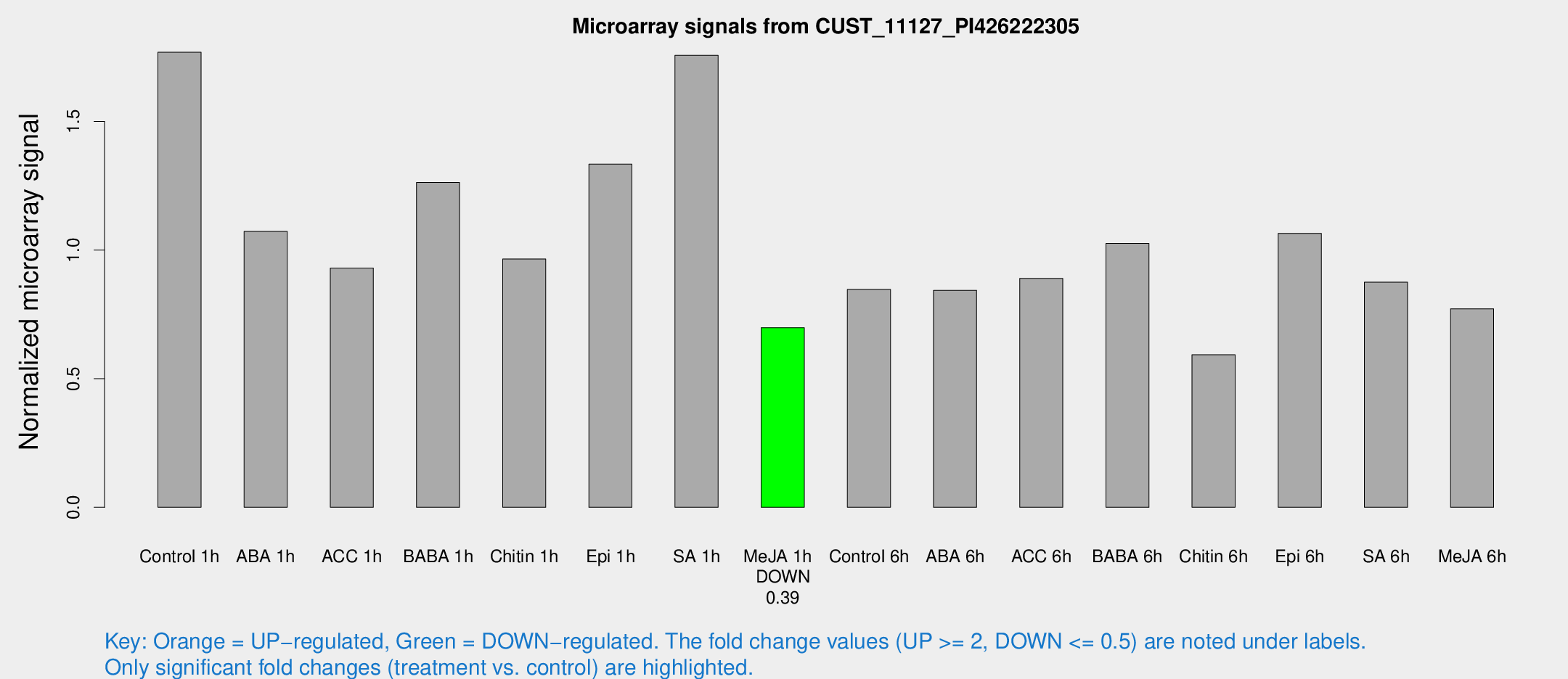
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 220.192 | 31.789 | 1.76959 | 0.135588 |
| ABA 1h | 118.743 | 19.0617 | 1.07277 | 0.136979 |
| ACC 1h | 130.627 | 37.2339 | 0.930587 | 0.280935 |
| BABA 1h | 154.049 | 27.0714 | 1.26314 | 0.116158 |
| Chitin 1h | 106.478 | 7.89751 | 0.965321 | 0.0627723 |
| Epi 1h | 142.121 | 12.7745 | 1.3343 | 0.10367 |
| SA 1h | 220.48 | 13.1522 | 1.7578 | 0.104784 |
| Me-JA 1h | 71.6422 | 12.2943 | 0.698537 | 0.100257 |
| Control 6h | 112.762 | 29.2979 | 0.846893 | 0.185286 |
| ABA 6h | 114.765 | 26.0744 | 0.843808 | 0.138102 |
| ACC 6h | 124.816 | 8.27331 | 0.889954 | 0.0830938 |
| BABA 6h | 143.271 | 20.2561 | 1.0264 | 0.114321 |
| Chitin 6h | 78.2221 | 9.70367 | 0.593201 | 0.0638832 |
| Epi 6h | 147.347 | 10.8822 | 1.06531 | 0.0764141 |
| SA 6h | 108.898 | 19.5604 | 0.87546 | 0.107989 |
| Me-JA 6h | 98.0667 | 21.0436 | 0.771952 | 0.142127 |
Source Transcript PGSC0003DMT400078452 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G25390.1 | +3 | 9e-128 | 387 | 203/314 (65%) | Protein kinase superfamily protein | chr1:8906640-8908800 REVERSE LENGTH=629 |
