Probe CUST_10595_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10595_PI426222305 | JHI_St_60k_v1 | DMT400031994 | GATGCTGAAATTGTATATGCTGCAACAGAGAAAGCAGAACGCGAATTGAAGTGGAAGGGG |
All Microarray Probes Designed to Gene DMG400012274
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10473_PI426222305 | JHI_St_60k_v1 | DMT400031995 | ACGTCCTACCTTTAAATTCTGAATGCGCCTCTAAACACTCCGGTTTATGTTGCTTGCAGG |
| CUST_10595_PI426222305 | JHI_St_60k_v1 | DMT400031994 | GATGCTGAAATTGTATATGCTGCAACAGAGAAAGCAGAACGCGAATTGAAGTGGAAGGGG |
Microarray Signals from CUST_10595_PI426222305
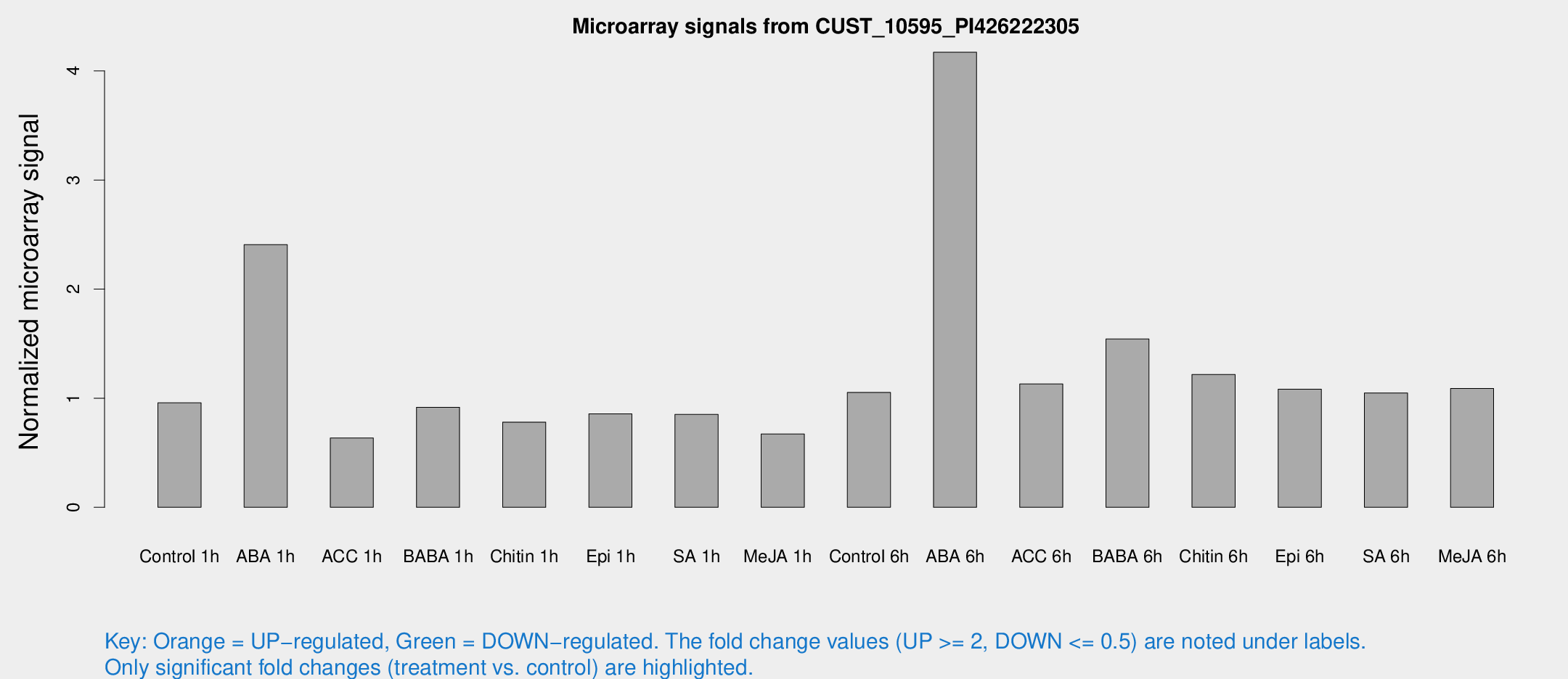
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 346.52 | 48.2991 | 0.957426 | 0.067987 |
| ABA 1h | 805.274 | 190.732 | 2.40829 | 0.465522 |
| ACC 1h | 274.132 | 90.3903 | 0.6354 | 0.264109 |
| BABA 1h | 336.163 | 79.0417 | 0.916951 | 0.159496 |
| Chitin 1h | 252.682 | 30.0334 | 0.78051 | 0.0667046 |
| Epi 1h | 264.856 | 20.4015 | 0.856551 | 0.0503752 |
| SA 1h | 339.099 | 101.057 | 0.851359 | 0.206847 |
| Me-JA 1h | 202.784 | 41.5285 | 0.6716 | 0.077256 |
| Control 6h | 418.676 | 131.318 | 1.05319 | 0.29639 |
| ABA 6h | 1732.07 | 466.072 | 4.17137 | 1.23382 |
| ACC 6h | 465.703 | 48.8058 | 1.13072 | 0.0658518 |
| BABA 6h | 630.542 | 102.562 | 1.54275 | 0.199638 |
| Chitin 6h | 460.693 | 26.8349 | 1.21749 | 0.0708605 |
| Epi 6h | 443.345 | 65.0548 | 1.08307 | 0.129106 |
| SA 6h | 395.27 | 101.854 | 1.04836 | 0.181423 |
| Me-JA 6h | 418.231 | 115.516 | 1.08866 | 0.233965 |
Source Transcript PGSC0003DMT400031994 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G10960.1 | +1 | 0.0 | 554 | 278/349 (80%) | UDP-D-glucose/UDP-D-galactose 4-epimerase 5 | chr4:6716083-6718472 REVERSE LENGTH=351 |
