Probe CUST_10030_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10030_PI426222305 | JHI_St_60k_v1 | DMT400028157 | GGCAAAGGAATACCTAATAGTGTGTCAATATAGTACTTTCATTTAAATCTCCACATCGTC |
All Microarray Probes Designed to Gene DMG400010859
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10030_PI426222305 | JHI_St_60k_v1 | DMT400028157 | GGCAAAGGAATACCTAATAGTGTGTCAATATAGTACTTTCATTTAAATCTCCACATCGTC |
| CUST_10050_PI426222305 | JHI_St_60k_v1 | DMT400028158 | GGCAAAGGAATACCTAATAGTGTGTCAATATAGTACTTTCATTTAAATCTCCACATCGTC |
Microarray Signals from CUST_10030_PI426222305
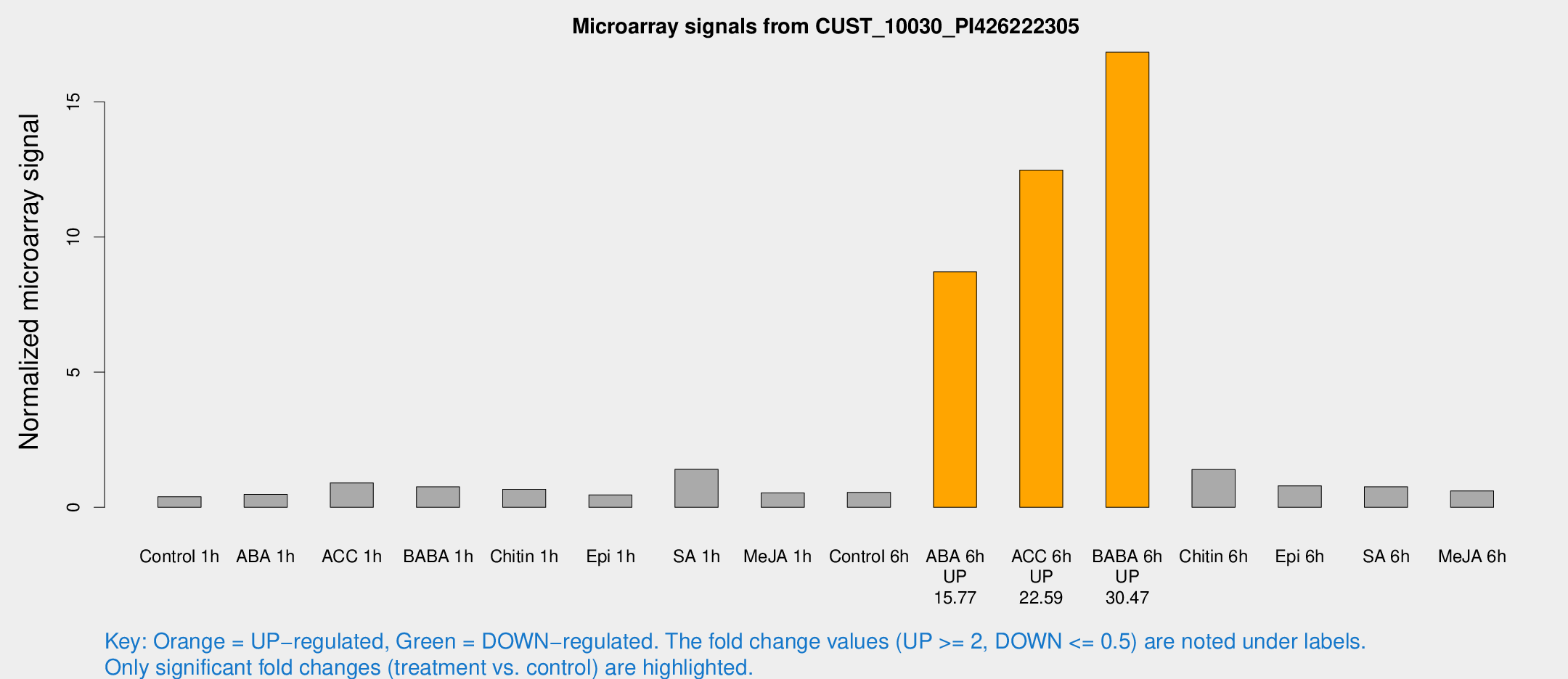
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.5754 | 4.78544 | 0.389028 | 0.177698 |
| ABA 1h | 23.4608 | 17.5655 | 0.479334 | 0.614919 |
| ACC 1h | 31.461 | 9.95963 | 0.904518 | 0.297253 |
| BABA 1h | 32.0649 | 14.0028 | 0.760104 | 0.649867 |
| Chitin 1h | 22.3682 | 10.0132 | 0.667922 | 0.350875 |
| Epi 1h | 12.1413 | 3.04138 | 0.459341 | 0.119269 |
| SA 1h | 52.9095 | 25.1819 | 1.40273 | 0.612589 |
| Me-JA 1h | 13.4353 | 3.17352 | 0.533183 | 0.131726 |
| Control 6h | 21.1417 | 8.36108 | 0.552636 | 0.297849 |
| ABA 6h | 329.282 | 129.297 | 8.71254 | 3.68253 |
| ACC 6h | 427.047 | 24.947 | 12.4814 | 1.87607 |
| BABA 6h | 669.187 | 266.686 | 16.8403 | 8.04328 |
| Chitin 6h | 49.2155 | 16.6402 | 1.39769 | 0.419032 |
| Epi 6h | 34.6138 | 13.5452 | 0.792431 | 0.503148 |
| SA 6h | 30.2682 | 14.53 | 0.762363 | 0.431148 |
| Me-JA 6h | 25.1727 | 11.4731 | 0.607046 | 0.44566 |
Source Transcript PGSC0003DMT400028157 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G55020.1 | +3 | 0.0 | 934 | 441/620 (71%) | lipoxygenase 1 | chr1:20525798-20530143 FORWARD LENGTH=859 |
