Probe CUST_9845_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9845_PI426222305 | JHI_St_60k_v1 | DMT400059525 | TATGTGATCCGTGTTTGCTCGCTCAAAAATGCTATATGTTATATGTTAAGTTGCACACAT |
All Microarray Probes Designed to Gene DMG400023125
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9845_PI426222305 | JHI_St_60k_v1 | DMT400059525 | TATGTGATCCGTGTTTGCTCGCTCAAAAATGCTATATGTTATATGTTAAGTTGCACACAT |
Microarray Signals from CUST_9845_PI426222305
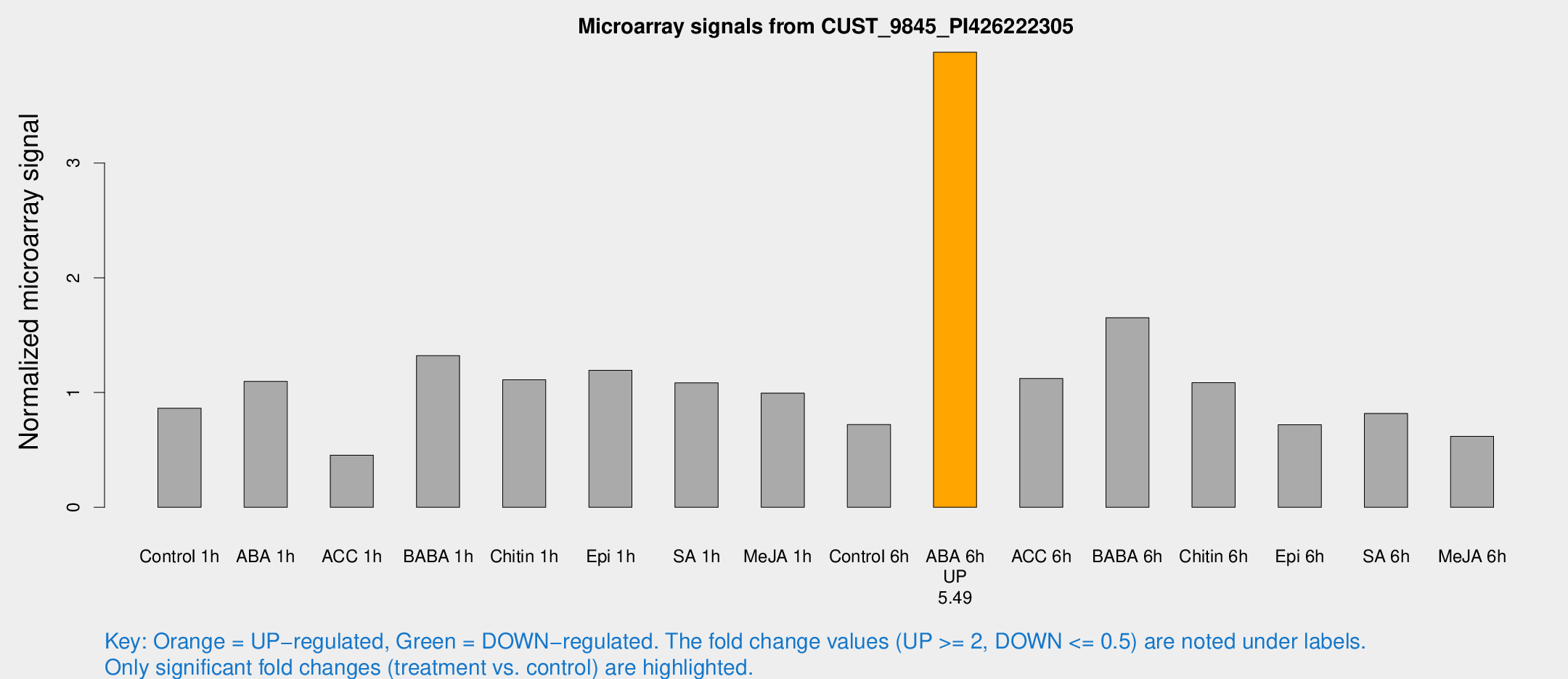
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1497.69 | 174.776 | 0.863346 | 0.161103 |
| ABA 1h | 1846.85 | 565.264 | 1.09736 | 0.313426 |
| ACC 1h | 1123.33 | 474.077 | 0.454058 | 0.369106 |
| BABA 1h | 2520.46 | 789.468 | 1.32178 | 0.443663 |
| Chitin 1h | 1763.18 | 295.507 | 1.11156 | 0.207215 |
| Epi 1h | 1792.96 | 216.11 | 1.1929 | 0.141787 |
| SA 1h | 2049.74 | 573.023 | 1.0853 | 0.23029 |
| Me-JA 1h | 1442.73 | 269.459 | 0.995116 | 0.19367 |
| Control 6h | 1270.96 | 220.232 | 0.721661 | 0.117018 |
| ABA 6h | 7364.23 | 1117.34 | 3.96522 | 0.36686 |
| ACC 6h | 2325.97 | 555.01 | 1.12311 | 0.113553 |
| BABA 6h | 3523.73 | 1122.93 | 1.65144 | 0.523564 |
| Chitin 6h | 1982.71 | 119.761 | 1.08588 | 0.0627261 |
| Epi 6h | 1393.38 | 88.492 | 0.720101 | 0.0554521 |
| SA 6h | 1462.29 | 314.643 | 0.817998 | 0.106478 |
| Me-JA 6h | 1320.66 | 565.713 | 0.619082 | 0.32444 |
Source Transcript PGSC0003DMT400059525 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G18210.1 | +2 | 8e-11 | 59 | 26/67 (39%) | Calcium-binding EF-hand family protein | chr1:6268273-6268785 REVERSE LENGTH=170 |
