Probe CUST_9427_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9427_PI426222305 | JHI_St_60k_v1 | DMT400023603 | GTTGCTTCCTCAAATATTGTTTCCTTTGCAACAAGACTTTGAGACTCGACAAAGAAGTTT |
All Microarray Probes Designed to Gene DMG400009139
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9423_PI426222305 | JHI_St_60k_v1 | DMT400023604 | TGTCCCATACCGTATGGGATTAAACAATGTGCTTAGTCAAAATCCTGACTTCATAAATTA |
| CUST_9427_PI426222305 | JHI_St_60k_v1 | DMT400023603 | GTTGCTTCCTCAAATATTGTTTCCTTTGCAACAAGACTTTGAGACTCGACAAAGAAGTTT |
Microarray Signals from CUST_9427_PI426222305
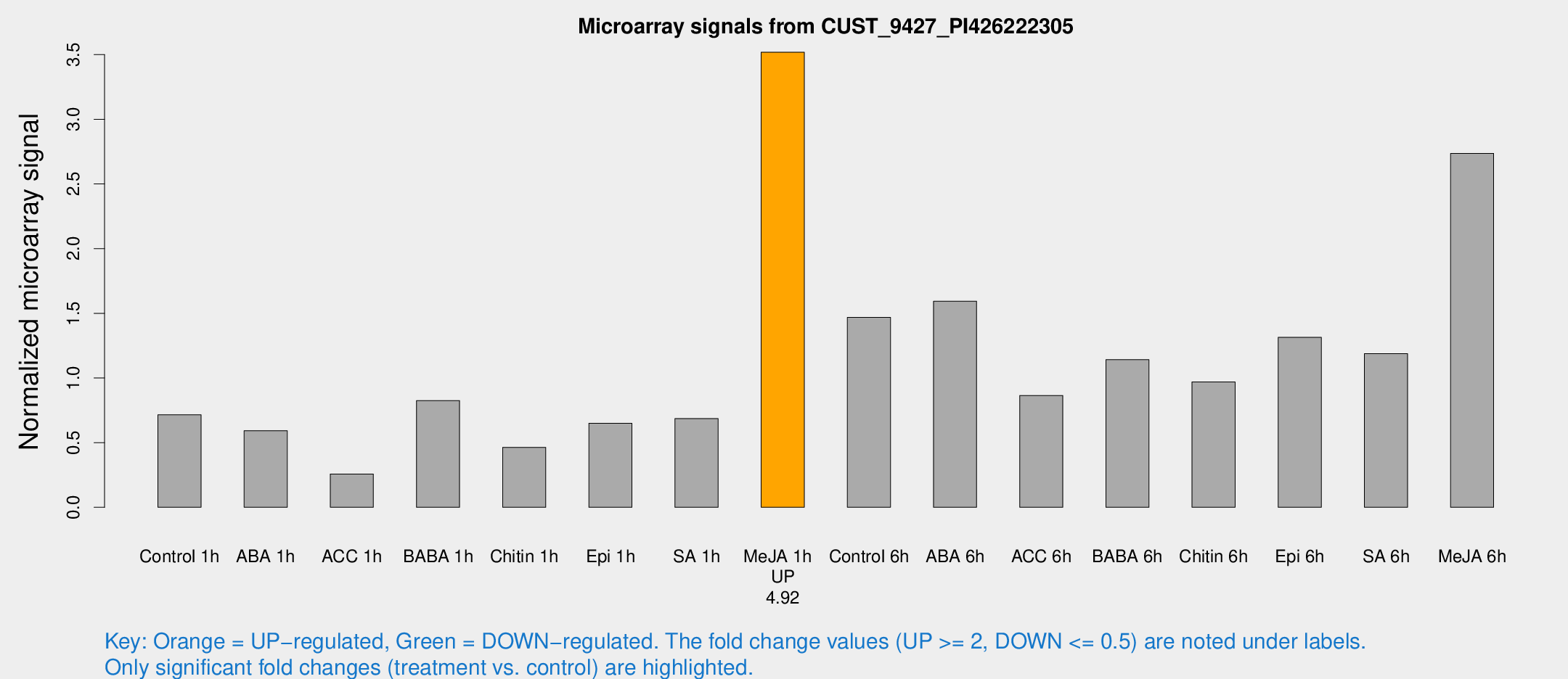
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 254.403 | 33.1498 | 0.7154 | 0.120822 |
| ABA 1h | 210.489 | 78.4503 | 0.592627 | 0.193357 |
| ACC 1h | 103.9 | 32.0621 | 0.257648 | 0.0796971 |
| BABA 1h | 290.158 | 57.0167 | 0.82506 | 0.0966163 |
| Chitin 1h | 146.505 | 11.9431 | 0.463548 | 0.0631459 |
| Epi 1h | 227.463 | 71.8759 | 0.649909 | 0.295248 |
| SA 1h | 256.616 | 55.2703 | 0.685948 | 0.185778 |
| Me-JA 1h | 1020.06 | 129.3 | 3.51864 | 0.648321 |
| Control 6h | 527.949 | 89.1953 | 1.4686 | 0.148314 |
| ABA 6h | 592.294 | 34.4334 | 1.5936 | 0.0925158 |
| ACC 6h | 349.625 | 34.4083 | 0.864565 | 0.0759827 |
| BABA 6h | 446.491 | 26.0925 | 1.14143 | 0.0666366 |
| Chitin 6h | 360.821 | 21.239 | 0.969154 | 0.056947 |
| Epi 6h | 529.011 | 74.7011 | 1.31403 | 0.300272 |
| SA 6h | 417.107 | 53.041 | 1.18855 | 0.0846419 |
| Me-JA 6h | 978.641 | 158.177 | 2.73598 | 0.335015 |
Source Transcript PGSC0003DMT400023603 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G53903.1 | +3 | 7e-25 | 98 | 56/131 (43%) | Protein of unknown function (DUF581) | chr1:20132363-20132842 FORWARD LENGTH=126 |
