Probe CUST_9423_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9423_PI426222305 | JHI_St_60k_v1 | DMT400023604 | TGTCCCATACCGTATGGGATTAAACAATGTGCTTAGTCAAAATCCTGACTTCATAAATTA |
All Microarray Probes Designed to Gene DMG400009139
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9427_PI426222305 | JHI_St_60k_v1 | DMT400023603 | GTTGCTTCCTCAAATATTGTTTCCTTTGCAACAAGACTTTGAGACTCGACAAAGAAGTTT |
| CUST_9423_PI426222305 | JHI_St_60k_v1 | DMT400023604 | TGTCCCATACCGTATGGGATTAAACAATGTGCTTAGTCAAAATCCTGACTTCATAAATTA |
Microarray Signals from CUST_9423_PI426222305
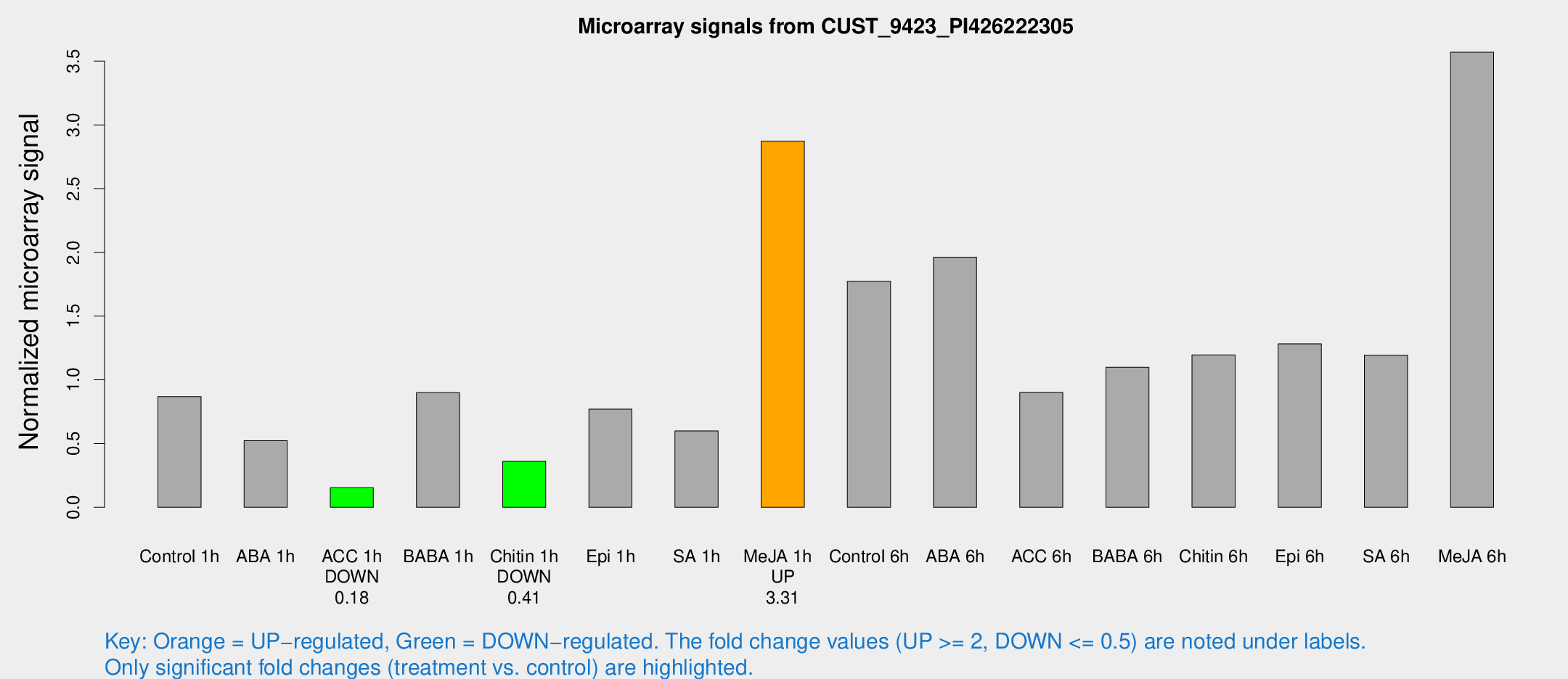
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 45.7139 | 4.02732 | 0.867845 | 0.0769522 |
| ABA 1h | 29.7835 | 13.1433 | 0.522714 | 0.227281 |
| ACC 1h | 8.89325 | 3.70287 | 0.153414 | 0.0730907 |
| BABA 1h | 47.0851 | 8.10097 | 0.898983 | 0.0860061 |
| Chitin 1h | 17.021 | 3.17717 | 0.360004 | 0.0667849 |
| Epi 1h | 40.1495 | 13.9406 | 0.770137 | 0.307039 |
| SA 1h | 33.3199 | 5.54999 | 0.598797 | 0.145289 |
| Me-JA 1h | 126.614 | 20.5827 | 2.87238 | 0.472359 |
| Control 6h | 96.1253 | 15.4111 | 1.77371 | 0.150344 |
| ABA 6h | 112.747 | 18.4093 | 1.96199 | 0.244523 |
| ACC 6h | 54.328 | 4.91122 | 0.901002 | 0.0944044 |
| BABA 6h | 67.2448 | 12.7359 | 1.09921 | 0.196353 |
| Chitin 6h | 67.7415 | 8.02309 | 1.19517 | 0.106566 |
| Epi 6h | 76.4906 | 6.73235 | 1.28278 | 0.163399 |
| SA 6h | 66.643 | 16.1362 | 1.19453 | 0.200102 |
| Me-JA 6h | 207.127 | 58.0589 | 3.56995 | 1.04523 |
Source Transcript PGSC0003DMT400023604 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G53903.1 | +1 | 2e-11 | 61 | 31/60 (52%) | Protein of unknown function (DUF581) | chr1:20132363-20132842 FORWARD LENGTH=126 |
