Probe CUST_9133_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9133_PI426222305 | JHI_St_60k_v1 | DMT400013037 | CTAAAGCAAGGGAGTTTAGTGTGCATTGAGCATATATAATTATCCAAGCTAATCTCTTTC |
All Microarray Probes Designed to Gene DMG400005082
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9133_PI426222305 | JHI_St_60k_v1 | DMT400013037 | CTAAAGCAAGGGAGTTTAGTGTGCATTGAGCATATATAATTATCCAAGCTAATCTCTTTC |
Microarray Signals from CUST_9133_PI426222305
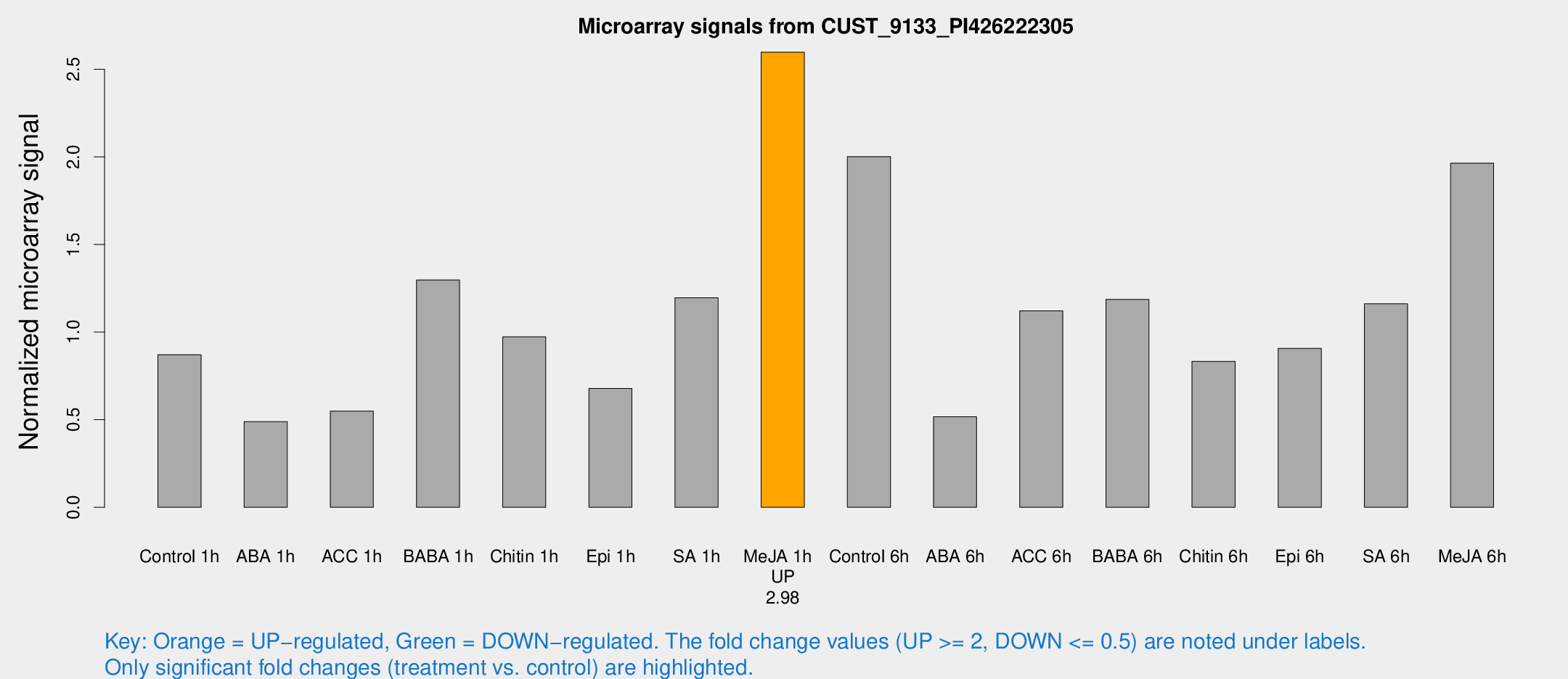
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 25.478 | 3.64439 | 0.870842 | 0.12548 |
| ABA 1h | 14.6406 | 4.94349 | 0.488852 | 0.227036 |
| ACC 1h | 18.0403 | 4.86409 | 0.548546 | 0.178188 |
| BABA 1h | 36.8351 | 4.37266 | 1.29717 | 0.249559 |
| Chitin 1h | 28.6951 | 8.40724 | 0.973176 | 0.32999 |
| Epi 1h | 17.2557 | 3.39775 | 0.67847 | 0.138076 |
| SA 1h | 38.9582 | 12.0053 | 1.19593 | 0.402594 |
| Me-JA 1h | 61.7701 | 4.94089 | 2.59739 | 0.206271 |
| Control 6h | 67.4588 | 21.8092 | 2.00159 | 0.691636 |
| ABA 6h | 18.0186 | 5.54699 | 0.516983 | 0.189711 |
| ACC 6h | 38.0429 | 4.96728 | 1.1214 | 0.139703 |
| BABA 6h | 39.1721 | 4.8987 | 1.18685 | 0.188242 |
| Chitin 6h | 26.3514 | 4.22242 | 0.832779 | 0.140095 |
| Epi 6h | 30.0575 | 4.371 | 0.907114 | 0.161258 |
| SA 6h | 35.4732 | 9.11172 | 1.16163 | 0.216195 |
| Me-JA 6h | 59.9674 | 14.6658 | 1.964 | 0.641015 |
Source Transcript PGSC0003DMT400013037 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G61820.1 | +1 | 0.0 | 538 | 264/454 (58%) | beta glucosidase 46 | chr1:22835452-22838444 FORWARD LENGTH=516 |
