Probe CUST_9119_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9119_PI426222305 | JHI_St_60k_v1 | DMT400058117 | TTCTTGACAAGCTTGATAATCACTATTTTAGCAGCAGTTTGGGATATGAAGCAGTTGCAT |
All Microarray Probes Designed to Gene DMG400022559
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9119_PI426222305 | JHI_St_60k_v1 | DMT400058117 | TTCTTGACAAGCTTGATAATCACTATTTTAGCAGCAGTTTGGGATATGAAGCAGTTGCAT |
Microarray Signals from CUST_9119_PI426222305
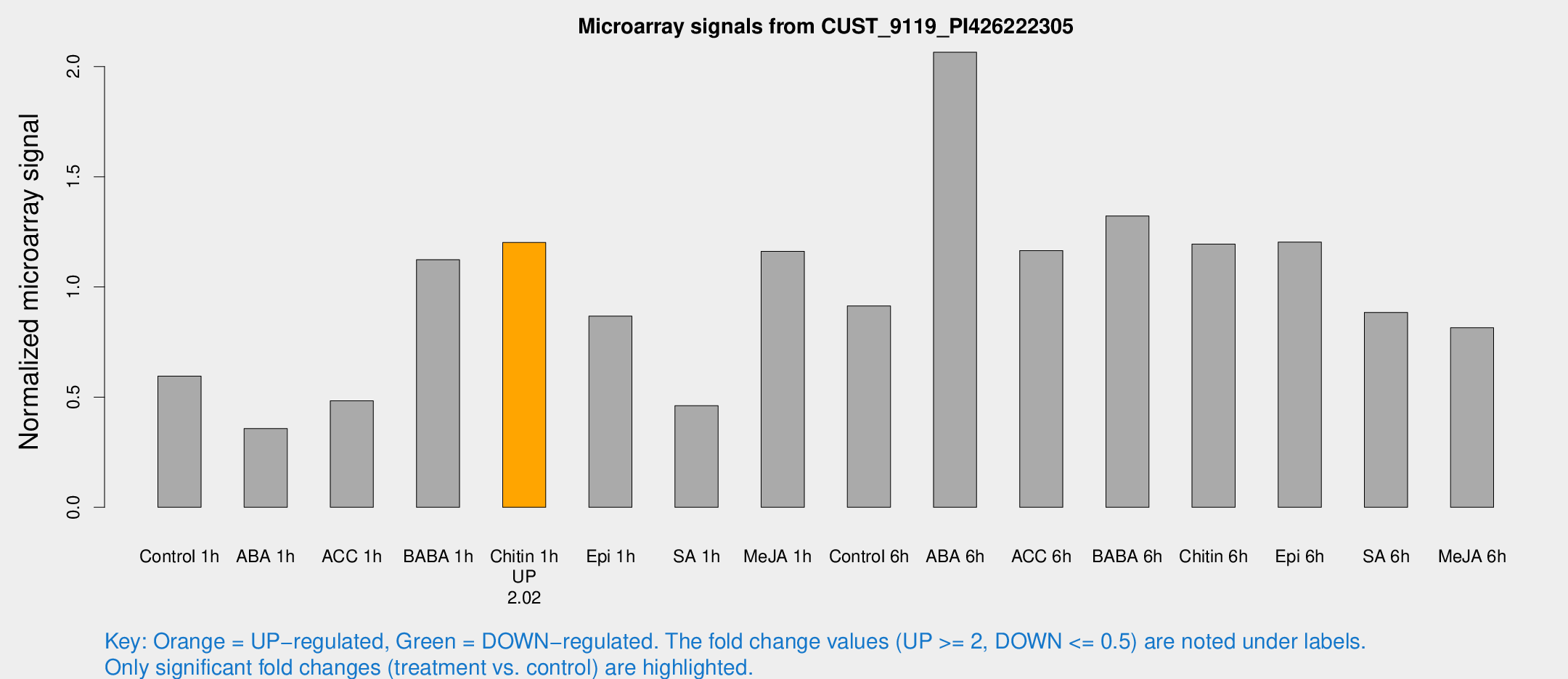
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.6509 | 3.73431 | 0.595428 | 0.166707 |
| ABA 1h | 7.27182 | 3.50929 | 0.357289 | 0.180056 |
| ACC 1h | 12.3389 | 4.92377 | 0.483018 | 0.224668 |
| BABA 1h | 24.9669 | 4.03997 | 1.12349 | 0.19034 |
| Chitin 1h | 24.9769 | 3.91103 | 1.20182 | 0.197102 |
| Epi 1h | 18.0164 | 4.09881 | 0.867488 | 0.207582 |
| SA 1h | 12.2608 | 4.54271 | 0.46119 | 0.178792 |
| Me-JA 1h | 21.5705 | 3.87521 | 1.16149 | 0.212609 |
| Control 6h | 28.7831 | 16.6092 | 0.913893 | 0.640267 |
| ABA 6h | 52.237 | 10.5716 | 2.06504 | 0.39151 |
| ACC 6h | 30.7152 | 4.87826 | 1.16505 | 0.208445 |
| BABA 6h | 34.7527 | 6.32284 | 1.32198 | 0.208318 |
| Chitin 6h | 29.1068 | 4.51581 | 1.19398 | 0.189346 |
| Epi 6h | 31.1633 | 4.77899 | 1.20372 | 0.187778 |
| SA 6h | 23.0971 | 7.5113 | 0.884481 | 0.292331 |
| Me-JA 6h | 20.6407 | 5.92891 | 0.814724 | 0.254535 |
Source Transcript PGSC0003DMT400058117 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G36010.1 | +1 | 2e-40 | 139 | 58/73 (79%) | Pathogenesis-related thaumatin superfamily protein | chr4:17039472-17040976 REVERSE LENGTH=301 |
