Probe CUST_9061_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9061_PI426222305 | JHI_St_60k_v1 | DMT400033379 | CCAGCTTCATTACAAGCATCAATTAAAGCTGCAACACCTGAAGAAACTATGAAGAATTTT |
All Microarray Probes Designed to Gene DMG400012822
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9061_PI426222305 | JHI_St_60k_v1 | DMT400033379 | CCAGCTTCATTACAAGCATCAATTAAAGCTGCAACACCTGAAGAAACTATGAAGAATTTT |
Microarray Signals from CUST_9061_PI426222305
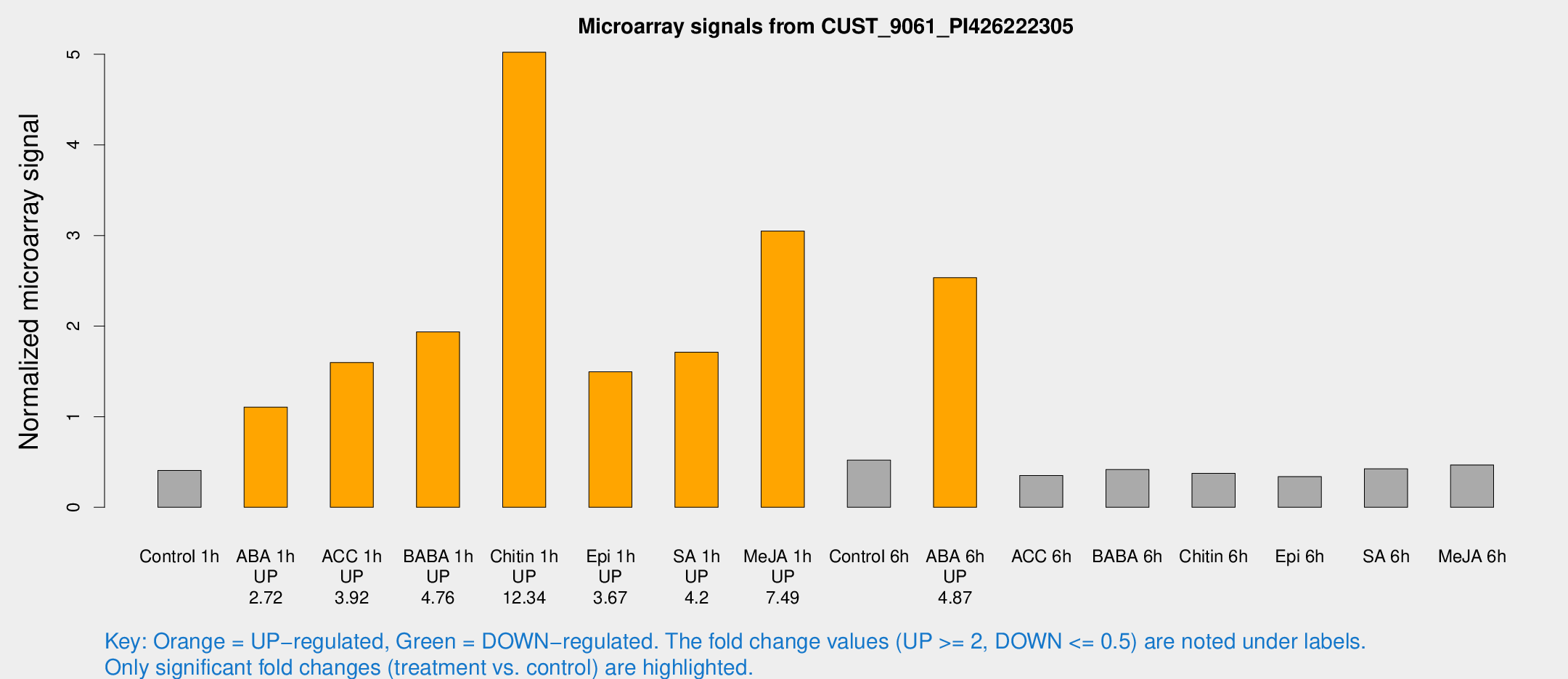
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.70714 | 3.73296 | 0.407116 | 0.161362 |
| ABA 1h | 23.4197 | 3.90491 | 1.10653 | 0.189052 |
| ACC 1h | 39.4145 | 5.6816 | 1.59745 | 0.367821 |
| BABA 1h | 47.2401 | 12.0212 | 1.93672 | 0.362207 |
| Chitin 1h | 122.715 | 48.7034 | 5.02268 | 2.03023 |
| Epi 1h | 31.3611 | 5.65145 | 1.49528 | 0.27254 |
| SA 1h | 46.7998 | 16.1109 | 1.71114 | 0.537094 |
| Me-JA 1h | 61.5856 | 12.8056 | 3.04886 | 0.562251 |
| Control 6h | 13.2324 | 3.8722 | 0.520541 | 0.176129 |
| ABA 6h | 63.7217 | 5.38325 | 2.53406 | 0.320136 |
| ACC 6h | 10.8286 | 4.8105 | 0.35168 | 0.175629 |
| BABA 6h | 13.6282 | 6.60927 | 0.416693 | 0.199176 |
| Chitin 6h | 9.68667 | 4.27903 | 0.374301 | 0.176944 |
| Epi 6h | 9.01687 | 4.59698 | 0.338118 | 0.174591 |
| SA 6h | 10.1659 | 4.10142 | 0.425894 | 0.183339 |
| Me-JA 6h | 14.7183 | 8.21867 | 0.467695 | 0.298541 |
Source Transcript PGSC0003DMT400033379 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G27450.1 | +3 | 3e-111 | 328 | 160/249 (64%) | Aluminium induced protein with YGL and LRDR motifs | chr4:13727665-13728683 REVERSE LENGTH=250 |
