Probe CUST_9025_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9025_PI426222305 | JHI_St_60k_v1 | DMT400061846 | CTCAGGATAATACTGTGGGCACGATAACTGATATACCAAATTCATTTTTGGTTCATTAGA |
All Microarray Probes Designed to Gene DMG400024063
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9025_PI426222305 | JHI_St_60k_v1 | DMT400061846 | CTCAGGATAATACTGTGGGCACGATAACTGATATACCAAATTCATTTTTGGTTCATTAGA |
| CUST_9059_PI426222305 | JHI_St_60k_v1 | DMT400061845 | CCTATTGCATATGCAAAATCTCTTGTGCCTCCTACAAGAACTATCTCTCTTCTAAGCTAA |
Microarray Signals from CUST_9025_PI426222305
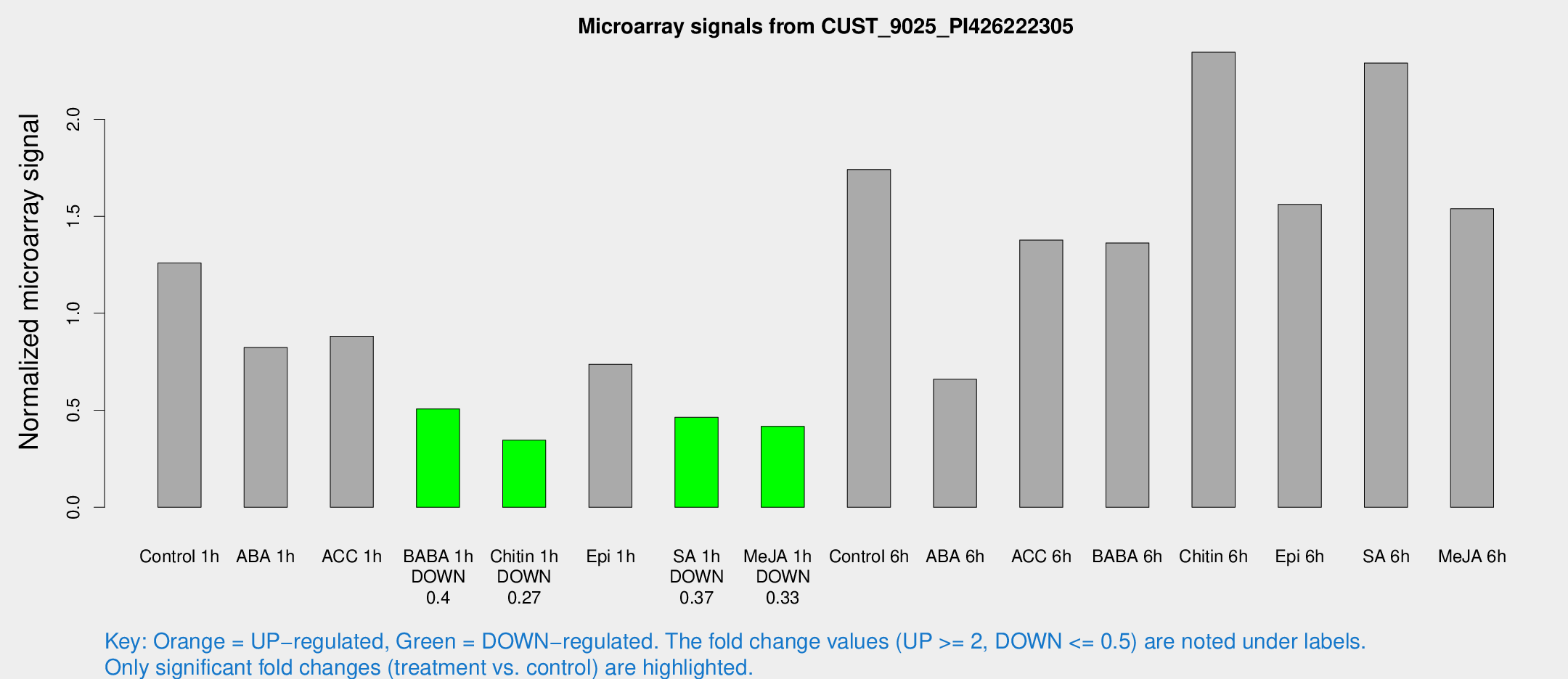
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 296.139 | 54.5015 | 1.25964 | 0.17844 |
| ABA 1h | 175.156 | 39.5385 | 0.823725 | 0.25054 |
| ACC 1h | 211.899 | 32.8269 | 0.882199 | 0.0844712 |
| BABA 1h | 113.511 | 15.2259 | 0.506993 | 0.0343898 |
| Chitin 1h | 70.9805 | 5.46757 | 0.346151 | 0.026729 |
| Epi 1h | 146.499 | 13.7028 | 0.737145 | 0.0699176 |
| SA 1h | 109.226 | 9.16113 | 0.463941 | 0.0427924 |
| Me-JA 1h | 78.1944 | 6.78883 | 0.417527 | 0.0397396 |
| Control 6h | 405.418 | 59.6946 | 1.74074 | 0.287394 |
| ABA 6h | 162.018 | 18.8448 | 0.660132 | 0.108409 |
| ACC 6h | 363.591 | 36.844 | 1.37759 | 0.0955747 |
| BABA 6h | 357.884 | 58.9579 | 1.36244 | 0.288271 |
| Chitin 6h | 583.447 | 94.4365 | 2.34551 | 0.308603 |
| Epi 6h | 408.051 | 56.5565 | 1.56152 | 0.123572 |
| SA 6h | 921.032 | 670.812 | 2.28988 | 2.26788 |
| Me-JA 6h | 352.735 | 35.8631 | 1.539 | 0.0904575 |
Source Transcript PGSC0003DMT400061846 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G17230.3 | +2 | 0.0 | 570 | 299/432 (69%) | PHYTOENE SYNTHASE | chr5:5659839-5662087 REVERSE LENGTH=437 |
