Probe CUST_8963_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8963_PI426222305 | JHI_St_60k_v1 | DMT400033390 | TAACGATCTCAAGCCAAAAGCTAATTTCGACCTTAATCTTCTCCCGGCACCGGAAGACAT |
All Microarray Probes Designed to Gene DMG400012828
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8931_PI426222305 | JHI_St_60k_v1 | DMT400033391 | AGGGGAAAACGATGACGTTCTGTTCATCGAGTTACTATGTGAATATCGAAAAATCTGAAC |
| CUST_8963_PI426222305 | JHI_St_60k_v1 | DMT400033390 | TAACGATCTCAAGCCAAAAGCTAATTTCGACCTTAATCTTCTCCCGGCACCGGAAGACAT |
Microarray Signals from CUST_8963_PI426222305
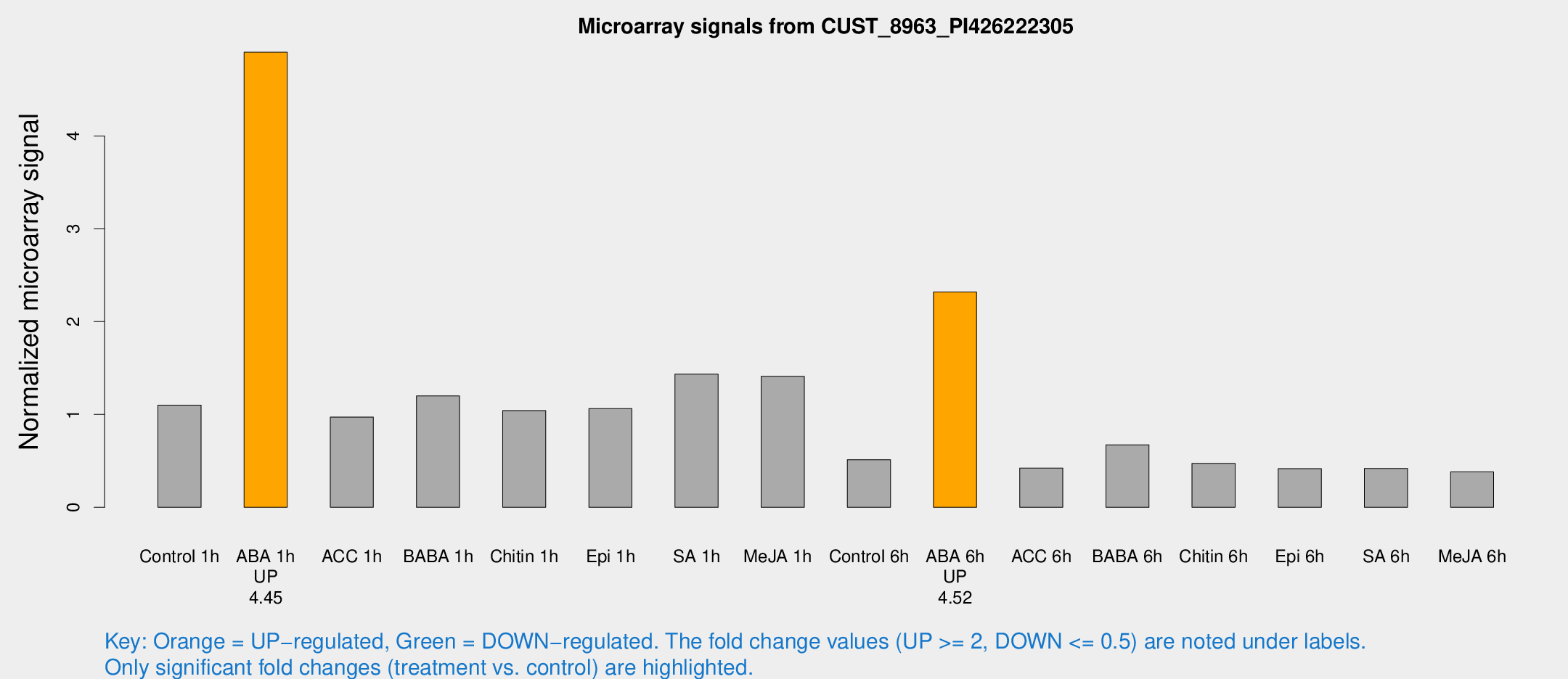
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 98.018 | 6.68156 | 1.10129 | 0.0749152 |
| ABA 1h | 398.692 | 73.1925 | 4.9018 | 0.655055 |
| ACC 1h | 114.155 | 49.1609 | 0.972086 | 0.544657 |
| BABA 1h | 104.031 | 13.1411 | 1.19986 | 0.0991329 |
| Chitin 1h | 83.6875 | 7.64073 | 1.04137 | 0.0744736 |
| Epi 1h | 82.4655 | 8.46227 | 1.0635 | 0.105617 |
| SA 1h | 132.717 | 17.7711 | 1.4341 | 0.218274 |
| Me-JA 1h | 102.239 | 6.93203 | 1.41007 | 0.0954711 |
| Control 6h | 47.999 | 10.0544 | 0.513286 | 0.0732552 |
| ABA 6h | 220.145 | 19.3676 | 2.31887 | 0.139446 |
| ACC 6h | 44.212 | 7.0084 | 0.423453 | 0.0560156 |
| BABA 6h | 67.6871 | 8.05964 | 0.673354 | 0.069183 |
| Chitin 6h | 45.1759 | 5.29622 | 0.472656 | 0.0709382 |
| Epi 6h | 41.5993 | 4.85358 | 0.415962 | 0.0491057 |
| SA 6h | 37.2486 | 4.42788 | 0.418763 | 0.0985437 |
| Me-JA 6h | 35.644 | 7.9322 | 0.38104 | 0.0656845 |
Source Transcript PGSC0003DMT400033390 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
