Probe CUST_8962_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8962_PI426222305 | JHI_St_60k_v1 | DMT400048075 | GCCAAAAATCAAAATGGCATCTCAATATTGGTGTTGTTTTGATCTGCACAAAGTGTTGTA |
All Microarray Probes Designed to Gene DMG400018679
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8962_PI426222305 | JHI_St_60k_v1 | DMT400048075 | GCCAAAAATCAAAATGGCATCTCAATATTGGTGTTGTTTTGATCTGCACAAAGTGTTGTA |
Microarray Signals from CUST_8962_PI426222305
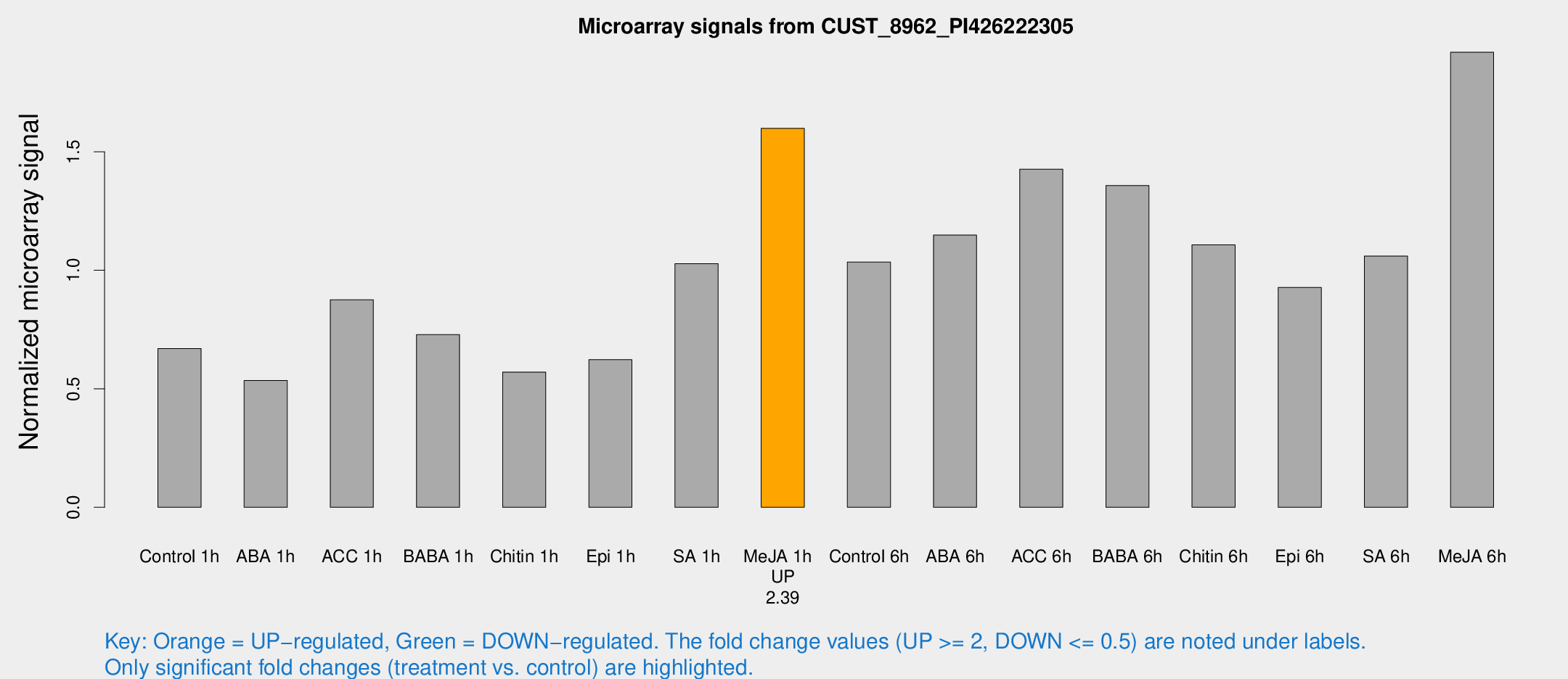
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 95.157 | 20.6672 | 0.669842 | 0.106778 |
| ABA 1h | 64.519 | 4.83424 | 0.535104 | 0.0401366 |
| ACC 1h | 124.559 | 17.3935 | 0.875502 | 0.0982932 |
| BABA 1h | 97.7652 | 14.1275 | 0.728394 | 0.0502909 |
| Chitin 1h | 70.9473 | 8.70063 | 0.5705 | 0.0454948 |
| Epi 1h | 73.4734 | 5.29047 | 0.622917 | 0.0449058 |
| SA 1h | 146.094 | 18.9279 | 1.02826 | 0.117467 |
| Me-JA 1h | 183.034 | 29.9442 | 1.59908 | 0.163959 |
| Control 6h | 156.505 | 44.1047 | 1.03474 | 0.267207 |
| ABA 6h | 177.648 | 47.9382 | 1.14868 | 0.236362 |
| ACC 6h | 223.678 | 15.1073 | 1.42674 | 0.158797 |
| BABA 6h | 213.302 | 37.4422 | 1.35756 | 0.190901 |
| Chitin 6h | 162.32 | 18.1988 | 1.10746 | 0.0870575 |
| Epi 6h | 143.751 | 13.9839 | 0.927784 | 0.160861 |
| SA 6h | 145.417 | 19.2017 | 1.05995 | 0.0669254 |
| Me-JA 6h | 261.186 | 16.9641 | 1.92015 | 0.294007 |
Source Transcript PGSC0003DMT400048075 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G76490.1 | +3 | 0.0 | 833 | 417/592 (70%) | hydroxy methylglutaryl CoA reductase 1 | chr1:28695801-28698206 FORWARD LENGTH=642 |
