Probe CUST_8855_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8855_PI426222305 | JHI_St_60k_v1 | DMT400086898 | ATTTCGGTGAATGTCGACCCACTTCTTAGGCCATCAATCTAGTGGTTTTGGTTTCGTGAC |
All Microarray Probes Designed to Gene DMG400036469
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8855_PI426222305 | JHI_St_60k_v1 | DMT400086898 | ATTTCGGTGAATGTCGACCCACTTCTTAGGCCATCAATCTAGTGGTTTTGGTTTCGTGAC |
Microarray Signals from CUST_8855_PI426222305
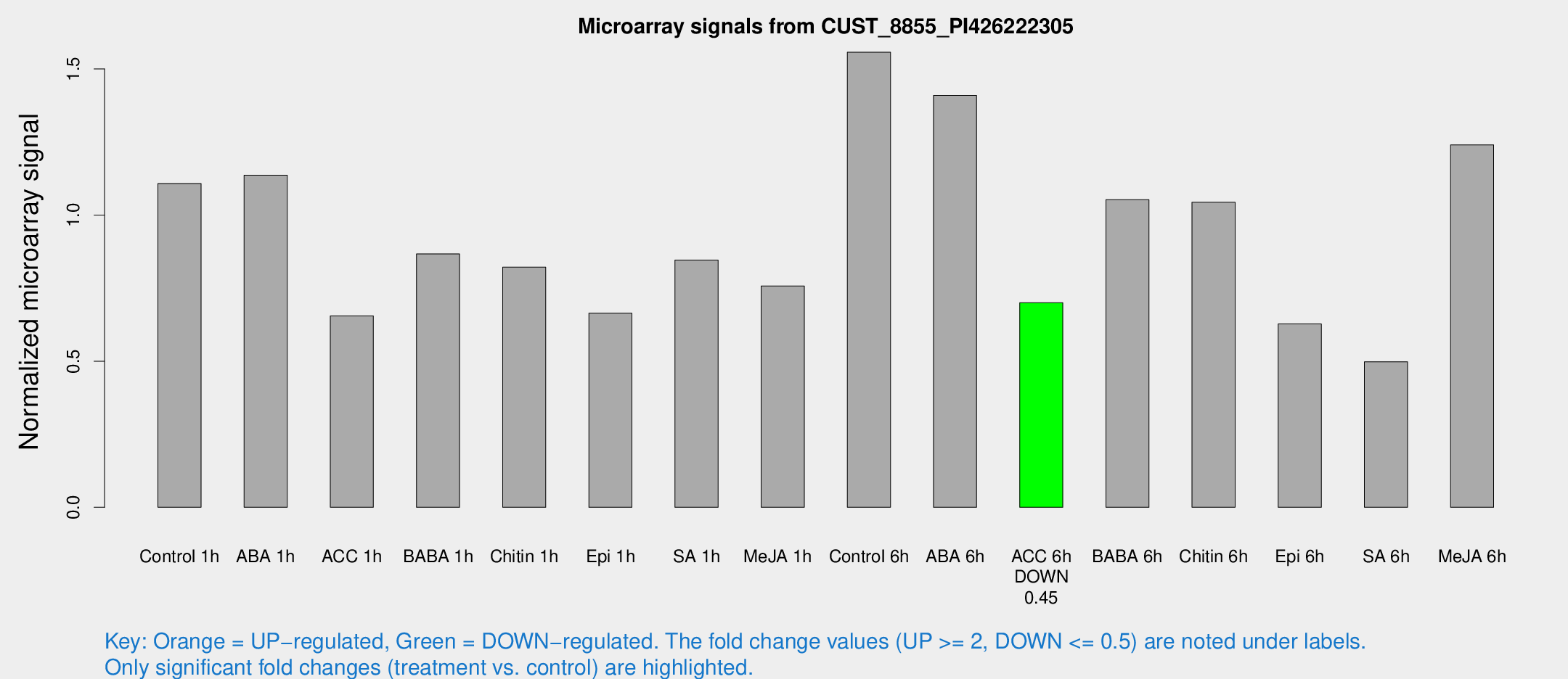
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 35.2736 | 4.04543 | 1.10762 | 0.153882 |
| ABA 1h | 32.7153 | 5.48999 | 1.13657 | 0.188357 |
| ACC 1h | 28.5793 | 13.1705 | 0.654949 | 0.421623 |
| BABA 1h | 28.1077 | 6.98591 | 0.866879 | 0.151327 |
| Chitin 1h | 23.454 | 3.87194 | 0.821568 | 0.137512 |
| Epi 1h | 23.6444 | 10.0567 | 0.664197 | 0.511064 |
| SA 1h | 28.099 | 4.01779 | 0.84622 | 0.176534 |
| Me-JA 1h | 26.4124 | 11.4654 | 0.757094 | 0.605751 |
| Control 6h | 50.0412 | 5.22007 | 1.55712 | 0.149509 |
| ABA 6h | 52.0132 | 13.7144 | 1.40917 | 0.421436 |
| ACC 6h | 26.2363 | 4.78127 | 0.699955 | 0.125537 |
| BABA 6h | 42.1645 | 12.4038 | 1.05271 | 0.390917 |
| Chitin 6h | 38.7864 | 10.4414 | 1.04375 | 0.3826 |
| Epi 6h | 26.6675 | 8.98304 | 0.627814 | 0.402146 |
| SA 6h | 31.0877 | 24.1692 | 0.497935 | 1.02443 |
| Me-JA 6h | 44.1471 | 13.7258 | 1.2401 | 0.371067 |
Source Transcript PGSC0003DMT400086898 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
