Probe CUST_8375_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8375_PI426222305 | JHI_St_60k_v1 | DMT400075209 | TCTATATAATGAAAGTTTTCTAGCACAAGATTGGGGAGGAAGTGACAGGGCCGGCTCTTG |
All Microarray Probes Designed to Gene DMG400029256
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7989_PI426222305 | JHI_St_60k_v1 | DMT400075210 | TGAATAGTAAACTCAATGAATTTCTTGAGAAAGCACGGCTTGGCTCTATCCATGAGCCAC |
| CUST_8375_PI426222305 | JHI_St_60k_v1 | DMT400075209 | TCTATATAATGAAAGTTTTCTAGCACAAGATTGGGGAGGAAGTGACAGGGCCGGCTCTTG |
Microarray Signals from CUST_8375_PI426222305
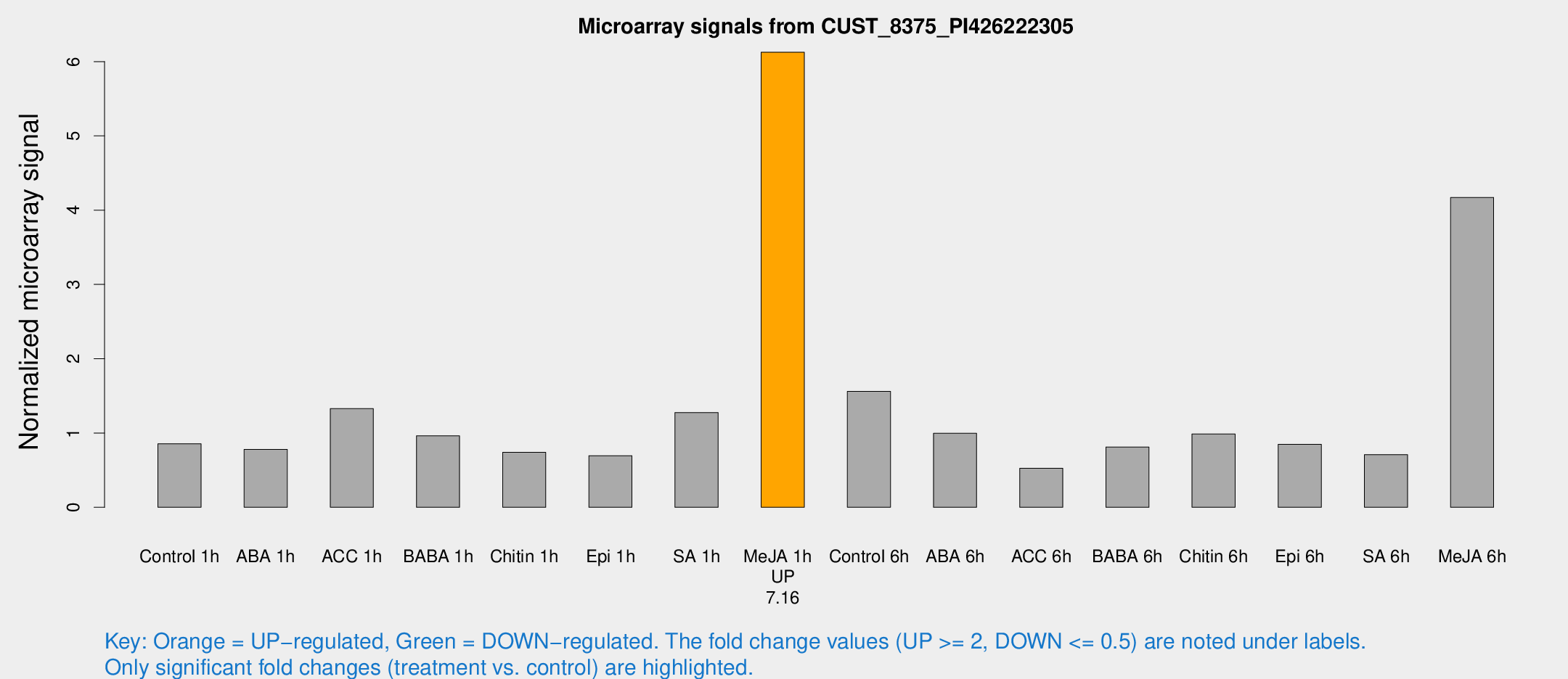
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 300.7 | 87.8835 | 0.855707 | 0.185645 |
| ABA 1h | 235.064 | 50.3361 | 0.780312 | 0.115391 |
| ACC 1h | 489.108 | 143.912 | 1.32785 | 0.513136 |
| BABA 1h | 316.47 | 71.7572 | 0.961827 | 0.220359 |
| Chitin 1h | 229.807 | 50.6739 | 0.740021 | 0.15184 |
| Epi 1h | 202.295 | 37.5984 | 0.695147 | 0.110475 |
| SA 1h | 545.978 | 239.559 | 1.27422 | 0.953334 |
| Me-JA 1h | 1637.21 | 136.88 | 6.12618 | 0.353967 |
| Control 6h | 650.467 | 321.863 | 1.56055 | 0.736442 |
| ABA 6h | 374.392 | 99.4782 | 0.996162 | 0.344583 |
| ACC 6h | 203.375 | 38.8179 | 0.525274 | 0.0445985 |
| BABA 6h | 304.181 | 50.9156 | 0.811317 | 0.179217 |
| Chitin 6h | 357.921 | 74.4132 | 0.986198 | 0.20909 |
| Epi 6h | 329.322 | 72.0775 | 0.847664 | 0.295914 |
| SA 6h | 311.261 | 124.409 | 0.709044 | 0.46665 |
| Me-JA 6h | 1361.52 | 120.961 | 4.17119 | 0.433693 |
Source Transcript PGSC0003DMT400075209 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G23970.1 | +1 | 5e-09 | 56 | 48/188 (26%) | HXXXD-type acyl-transferase family protein | chr5:8096326-8097612 FORWARD LENGTH=428 |
