Probe CUST_8374_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8374_PI426222305 | JHI_St_60k_v1 | DMT400075234 | TCAAGTTTGCTGGTTGTATGATGCAGAATGTGGCAATGGGATTTGTGTGCAAGTGGATTT |
All Microarray Probes Designed to Gene DMG400029263
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8374_PI426222305 | JHI_St_60k_v1 | DMT400075234 | TCAAGTTTGCTGGTTGTATGATGCAGAATGTGGCAATGGGATTTGTGTGCAAGTGGATTT |
Microarray Signals from CUST_8374_PI426222305
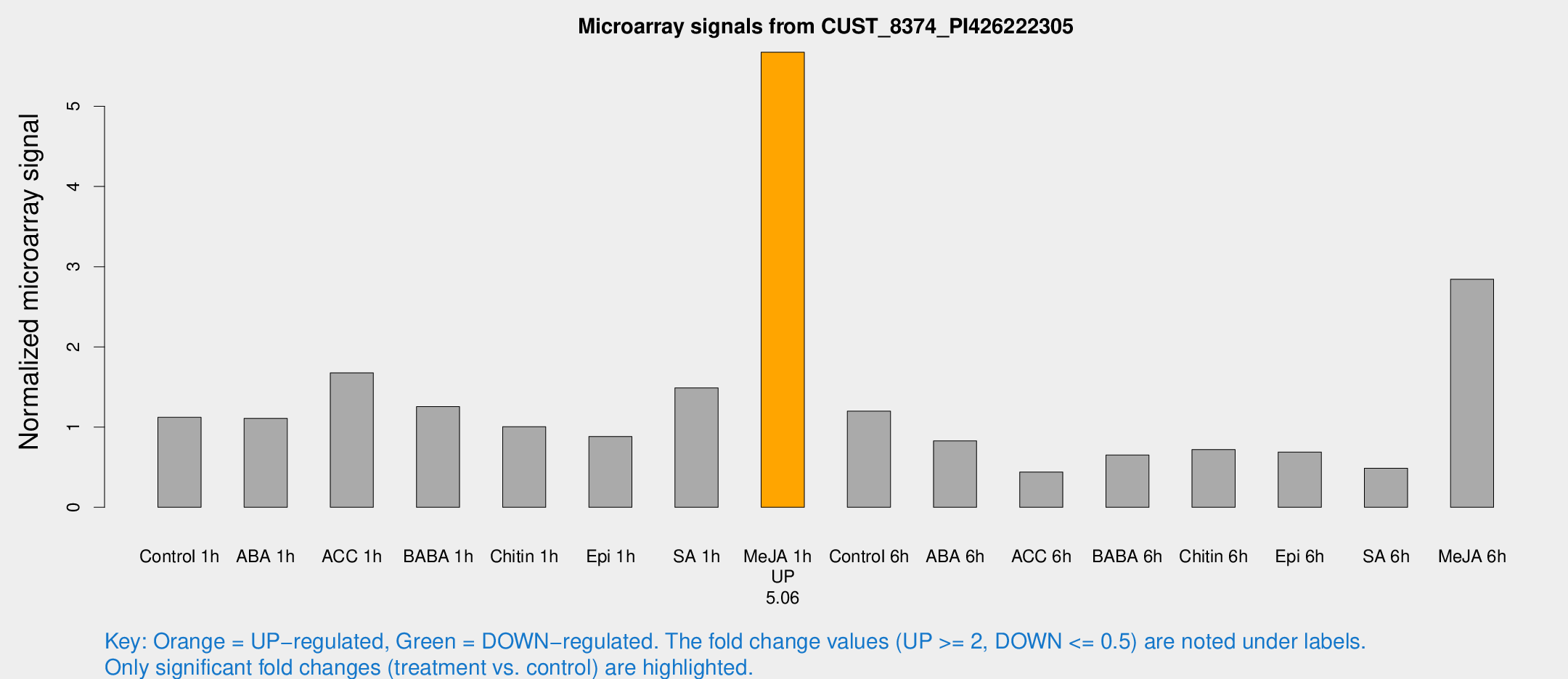
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9399.78 | 1215.68 | 1.12138 | 0.111278 |
| ABA 1h | 8384.66 | 1639.95 | 1.10924 | 0.15269 |
| ACC 1h | 14700.8 | 2872.84 | 1.67607 | 0.284237 |
| BABA 1h | 10086.1 | 1003.41 | 1.25534 | 0.229252 |
| Chitin 1h | 7865.8 | 1861.04 | 1.00497 | 0.224152 |
| Epi 1h | 6393.43 | 747.303 | 0.882842 | 0.102402 |
| SA 1h | 17866 | 9495.5 | 1.489 | 1.40126 |
| Me-JA 1h | 38477.3 | 3292.64 | 5.67395 | 0.820719 |
| Control 6h | 11770.8 | 4285.55 | 1.19876 | 0.45773 |
| ABA 6h | 7837.21 | 2276.5 | 0.827565 | 0.279746 |
| ACC 6h | 4379.98 | 958.325 | 0.439715 | 0.0739383 |
| BABA 6h | 6070.75 | 615.498 | 0.650821 | 0.0937408 |
| Chitin 6h | 6637.64 | 1530.53 | 0.720399 | 0.163082 |
| Epi 6h | 7030.91 | 1876.61 | 0.689247 | 0.305624 |
| SA 6h | 4781.92 | 1684.09 | 0.485926 | 0.192664 |
| Me-JA 6h | 23391.4 | 1352.66 | 2.84381 | 0.164188 |
Source Transcript PGSC0003DMT400075234 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G26040.1 | +1 | 2e-11 | 62 | 50/158 (32%) | HXXXD-type acyl-transferase family protein | chr3:9519741-9521069 FORWARD LENGTH=442 |
