Probe CUST_8362_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8362_PI426222305 | JHI_St_60k_v1 | DMT400091896 | CGGTCAACAACTTTGACTTGAATATTCCGGTGTTACCAGATAATTGGCCGGGTTTTGCCT |
All Microarray Probes Designed to Gene DMG400041467
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8362_PI426222305 | JHI_St_60k_v1 | DMT400091896 | CGGTCAACAACTTTGACTTGAATATTCCGGTGTTACCAGATAATTGGCCGGGTTTTGCCT |
Microarray Signals from CUST_8362_PI426222305
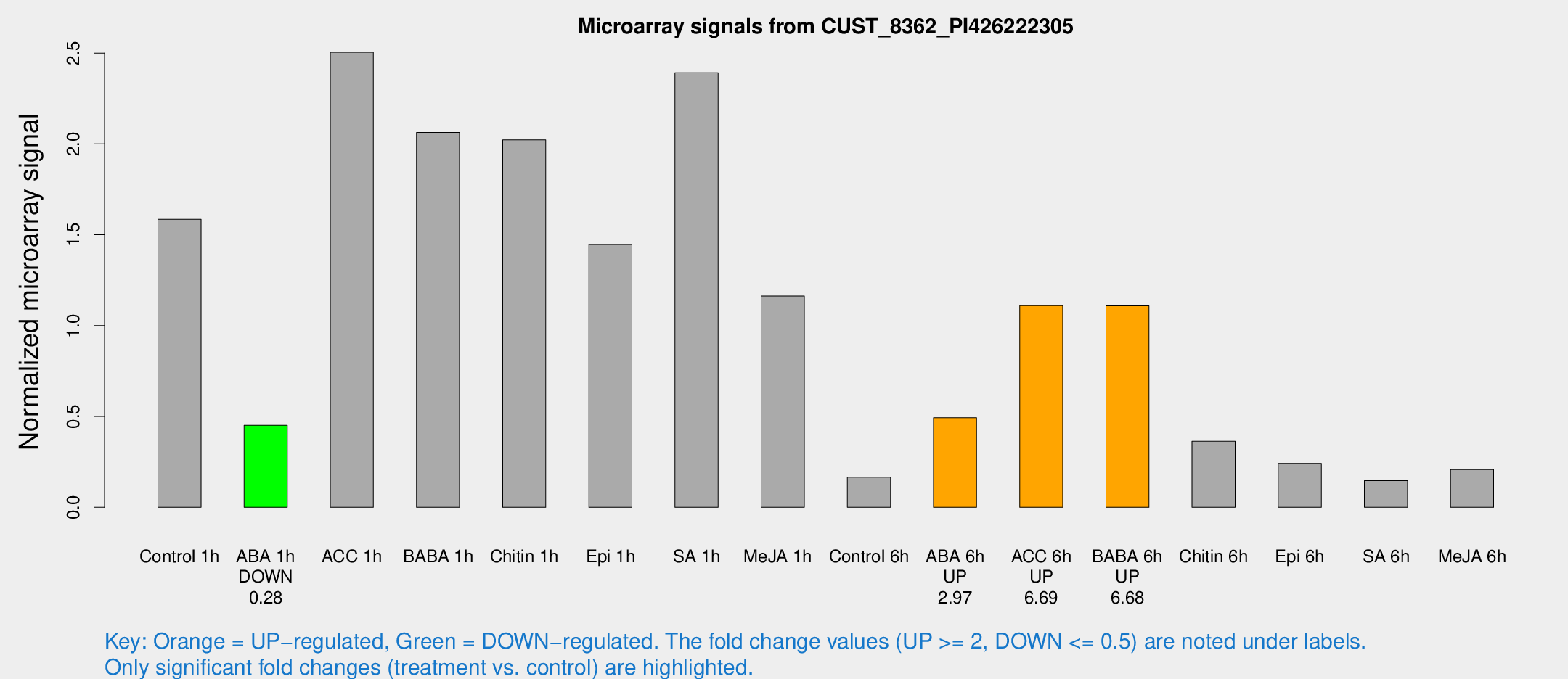
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1384.42 | 185.187 | 1.58562 | 0.145762 |
| ABA 1h | 343.249 | 20.05 | 0.451714 | 0.0263669 |
| ACC 1h | 2693.16 | 940.282 | 2.50453 | 1.20176 |
| BABA 1h | 1833.58 | 437.654 | 2.06348 | 0.371973 |
| Chitin 1h | 1564.16 | 90.5464 | 2.02235 | 0.241664 |
| Epi 1h | 1077.57 | 62.4547 | 1.44653 | 0.0836149 |
| SA 1h | 2125.79 | 187.198 | 2.39211 | 0.138152 |
| Me-JA 1h | 832.629 | 115.621 | 1.16282 | 0.0904785 |
| Control 6h | 159.838 | 48.2043 | 0.165995 | 0.0450742 |
| ABA 6h | 493.425 | 128.985 | 0.493304 | 0.145385 |
| ACC 6h | 1118.16 | 170.455 | 1.11069 | 0.06616 |
| BABA 6h | 1266.85 | 541.955 | 1.10908 | 0.456856 |
| Chitin 6h | 343.447 | 58.6515 | 0.363229 | 0.0722201 |
| Epi 6h | 248.115 | 53.7004 | 0.241953 | 0.0468829 |
| SA 6h | 126.929 | 16.2933 | 0.147172 | 0.0208561 |
| Me-JA 6h | 193.028 | 50.2252 | 0.207892 | 0.0468492 |
Source Transcript PGSC0003DMT400091896 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G27730.1 | +1 | 2e-41 | 142 | 95/198 (48%) | salt tolerance zinc finger | chr1:9648302-9648985 REVERSE LENGTH=227 |
