Probe CUST_8339_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8339_PI426222305 | JHI_St_60k_v1 | DMT400075404 | GAGTATAGACTTGAGGTCATGTCTTTTTAGGATTATTCCGGATGTACAATGCAATTTCAA |
All Microarray Probes Designed to Gene DMG400029328
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8339_PI426222305 | JHI_St_60k_v1 | DMT400075404 | GAGTATAGACTTGAGGTCATGTCTTTTTAGGATTATTCCGGATGTACAATGCAATTTCAA |
Microarray Signals from CUST_8339_PI426222305
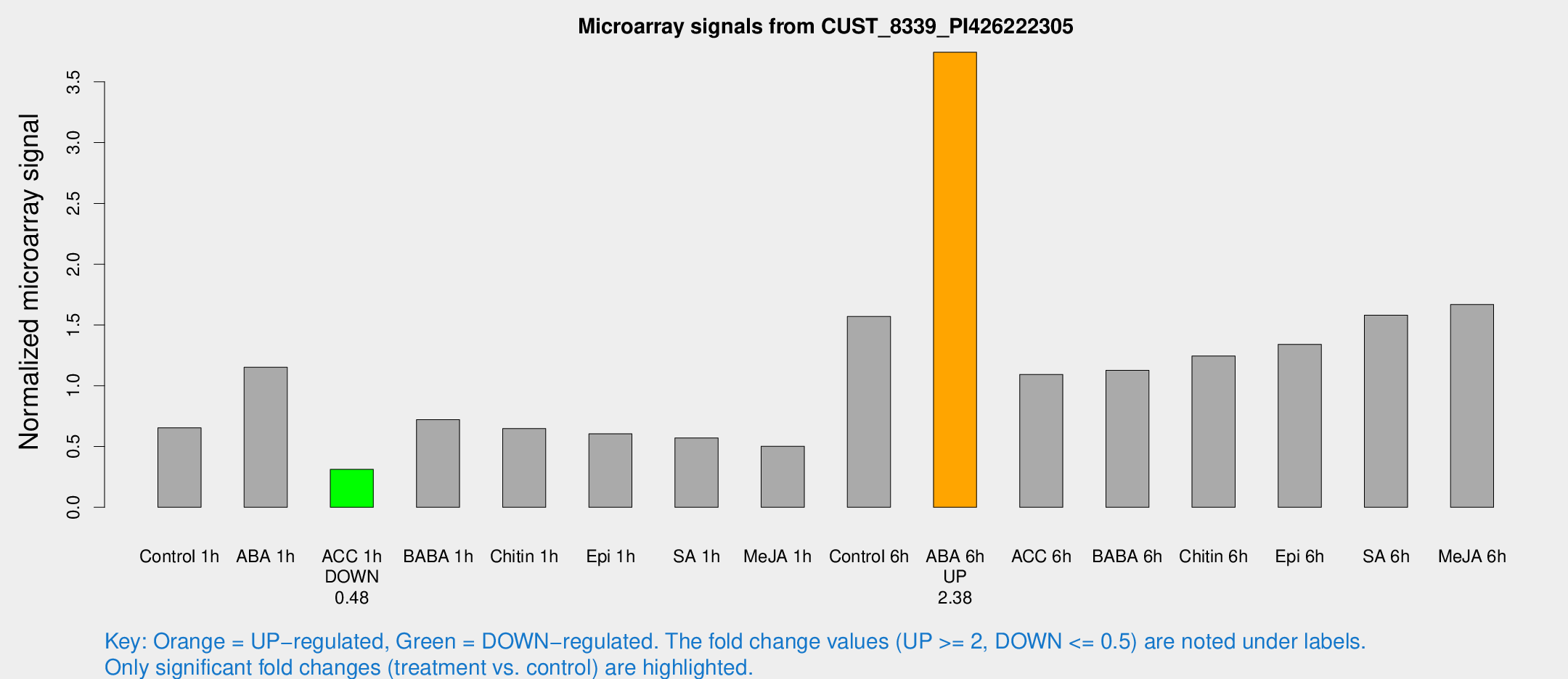
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 680.041 | 48.4742 | 0.654441 | 0.0758701 |
| ABA 1h | 1071.97 | 133.255 | 1.15238 | 0.103597 |
| ACC 1h | 335.595 | 37.1277 | 0.312286 | 0.0220647 |
| BABA 1h | 756.972 | 156.819 | 0.721234 | 0.0968649 |
| Chitin 1h | 603.126 | 35.0045 | 0.647578 | 0.0626487 |
| Epi 1h | 562.173 | 103.699 | 0.604184 | 0.114685 |
| SA 1h | 606.597 | 35.1698 | 0.570718 | 0.0330618 |
| Me-JA 1h | 427.103 | 35.4065 | 0.502511 | 0.0292171 |
| Control 6h | 1644.03 | 168.868 | 1.5699 | 0.0995855 |
| ABA 6h | 4155.59 | 424.21 | 3.74373 | 0.216163 |
| ACC 6h | 1316.54 | 155.598 | 1.09323 | 0.115443 |
| BABA 6h | 1313.41 | 116.285 | 1.12634 | 0.115194 |
| Chitin 6h | 1373.03 | 79.4996 | 1.24527 | 0.0719568 |
| Epi 6h | 1564.89 | 90.5562 | 1.34043 | 0.141155 |
| SA 6h | 1654.4 | 237.922 | 1.58101 | 0.0913378 |
| Me-JA 6h | 1766.4 | 280.738 | 1.66826 | 0.194016 |
Source Transcript PGSC0003DMT400075404 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G42200.1 | +3 | 4e-39 | 139 | 67/139 (48%) | RING/U-box superfamily protein | chr5:16860523-16861014 FORWARD LENGTH=163 |
