Probe CUST_8267_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8267_PI426222305 | JHI_St_60k_v1 | DMT400030193 | TCCTTCTCCTAGTTGGATCTTCAAGGCATGGGAAGCATCCGATGATGAAATAGTTGATGC |
All Microarray Probes Designed to Gene DMG400011555
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8267_PI426222305 | JHI_St_60k_v1 | DMT400030193 | TCCTTCTCCTAGTTGGATCTTCAAGGCATGGGAAGCATCCGATGATGAAATAGTTGATGC |
Microarray Signals from CUST_8267_PI426222305
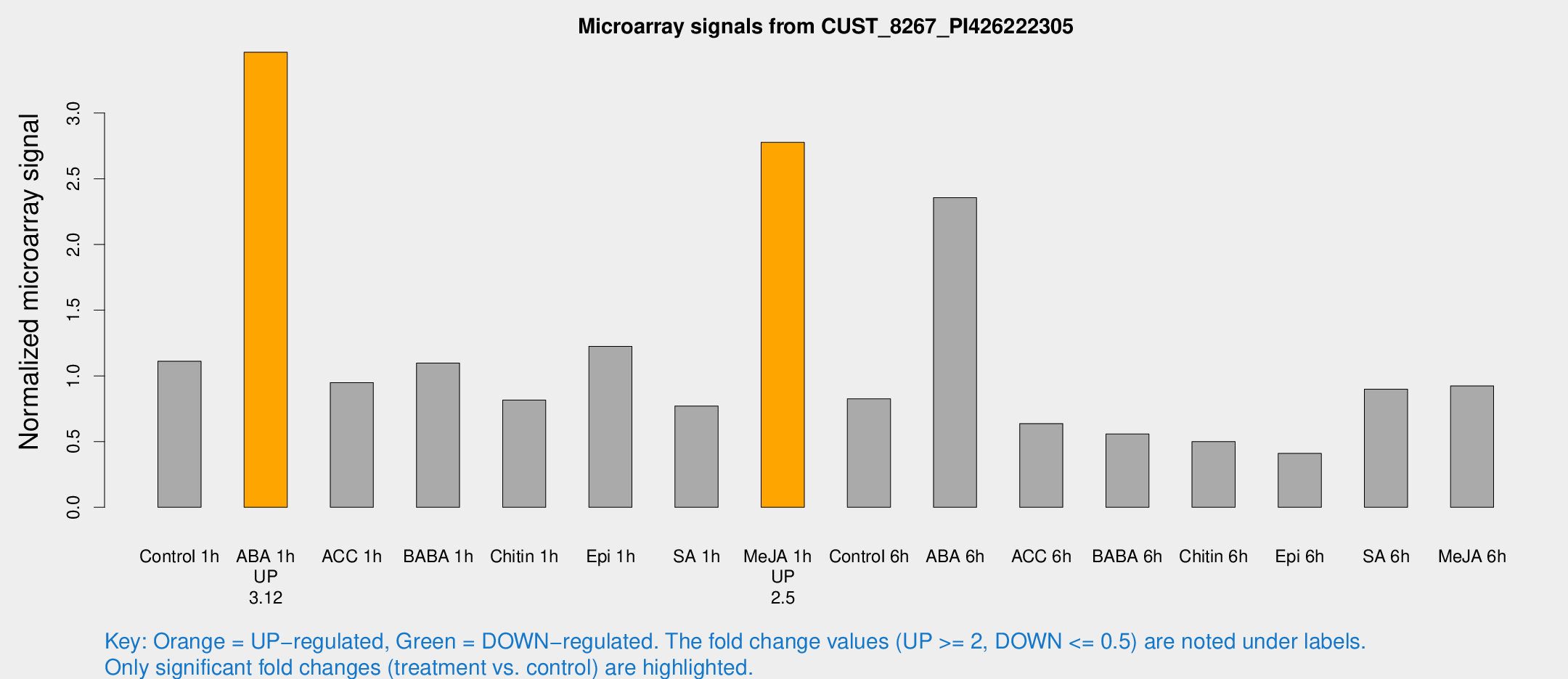
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 19.5699 | 3.41329 | 1.11159 | 0.200613 |
| ABA 1h | 53.3926 | 5.59915 | 3.46284 | 0.359014 |
| ACC 1h | 17.6515 | 3.93798 | 0.949042 | 0.221366 |
| BABA 1h | 19.1554 | 3.89597 | 1.09782 | 0.36546 |
| Chitin 1h | 17.0404 | 9.50861 | 0.815943 | 0.562294 |
| Epi 1h | 18.3083 | 3.32816 | 1.22474 | 0.223896 |
| SA 1h | 16.4773 | 6.36277 | 0.770679 | 0.471539 |
| Me-JA 1h | 39.4112 | 3.98769 | 2.77763 | 0.280122 |
| Control 6h | 14.5647 | 3.57408 | 0.825536 | 0.211868 |
| ABA 6h | 45.7668 | 10.5292 | 2.3566 | 0.53636 |
| ACC 6h | 14.3328 | 5.33661 | 0.63692 | 0.314311 |
| BABA 6h | 11.1855 | 3.83665 | 0.557914 | 0.216308 |
| Chitin 6h | 9.26133 | 3.86064 | 0.500144 | 0.213717 |
| Epi 6h | 8.26479 | 3.95899 | 0.410026 | 0.206233 |
| SA 6h | 17.9688 | 5.80303 | 0.898566 | 0.563448 |
| Me-JA 6h | 16.1839 | 3.54519 | 0.923432 | 0.209592 |
Source Transcript PGSC0003DMT400030193 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G10090.1 | +1 | 1e-119 | 384 | 195/394 (49%) | Early-responsive to dehydration stress protein (ERD4) | chr1:3290572-3295271 REVERSE LENGTH=762 |
