Probe CUST_8126_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8126_PI426222305 | JHI_St_60k_v1 | DMT400075430 | GGCTTTGAAGTTGAGTAATGCATGTGCATGTGTTGTGTTATGTTGTTGAACACATAATTA |
All Microarray Probes Designed to Gene DMG400029341
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8126_PI426222305 | JHI_St_60k_v1 | DMT400075430 | GGCTTTGAAGTTGAGTAATGCATGTGCATGTGTTGTGTTATGTTGTTGAACACATAATTA |
| CUST_8363_PI426222305 | JHI_St_60k_v1 | DMT400075429 | GAGGTCTTGATTAGTTCGAACATATTGTGGTAAGATATTCATGTAAGATAGACTATGCTG |
Microarray Signals from CUST_8126_PI426222305
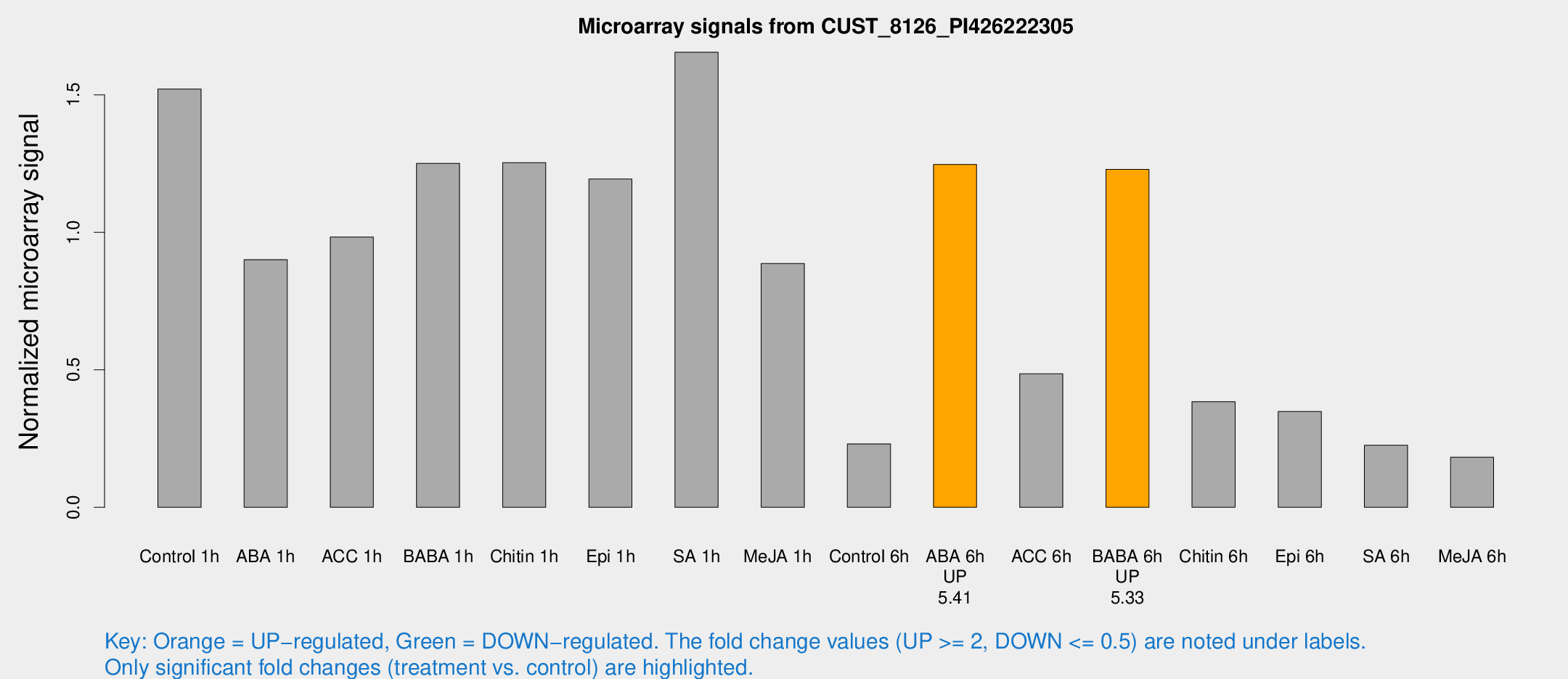
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 364.779 | 46.2211 | 1.52091 | 0.121696 |
| ABA 1h | 193.782 | 34.2242 | 0.900651 | 0.0924835 |
| ACC 1h | 279.824 | 91.5505 | 0.982804 | 0.388968 |
| BABA 1h | 296.634 | 55.1173 | 1.25075 | 0.131136 |
| Chitin 1h | 267.589 | 19.0087 | 1.2533 | 0.0950165 |
| Epi 1h | 245.702 | 20.5125 | 1.19367 | 0.071293 |
| SA 1h | 406.914 | 45.5366 | 1.65488 | 0.25369 |
| Me-JA 1h | 172.122 | 13.625 | 0.886531 | 0.101331 |
| Control 6h | 64.2381 | 26.2851 | 0.230615 | 0.0866566 |
| ABA 6h | 325.231 | 64.6673 | 1.24682 | 0.173445 |
| ACC 6h | 132.589 | 12.1275 | 0.485358 | 0.0933895 |
| BABA 6h | 343.569 | 84.1026 | 1.22916 | 0.247953 |
| Chitin 6h | 98.0798 | 12.1437 | 0.384113 | 0.0554922 |
| Epi 6h | 106.071 | 39.6102 | 0.348736 | 0.106259 |
| SA 6h | 54.9061 | 10.2132 | 0.225488 | 0.0231627 |
| Me-JA 6h | 54.597 | 25.8801 | 0.182394 | 0.0863577 |
Source Transcript PGSC0003DMT400075430 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
