Probe CUST_8014_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8014_PI426222305 | JHI_St_60k_v1 | DMT400039875 | TACGTGGTTTATCATGAGGCAATCTTGACGACAAAGGAGTACATGCAGTGTGTGACAGCT |
All Microarray Probes Designed to Gene DMG400015422
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8014_PI426222305 | JHI_St_60k_v1 | DMT400039875 | TACGTGGTTTATCATGAGGCAATCTTGACGACAAAGGAGTACATGCAGTGTGTGACAGCT |
| CUST_8354_PI426222305 | JHI_St_60k_v1 | DMT400039876 | TGACATTGTTGATACGAGTAATCTCAGACATGTTTGAAGACATGAGAGCAGCAATTCCAG |
Microarray Signals from CUST_8014_PI426222305
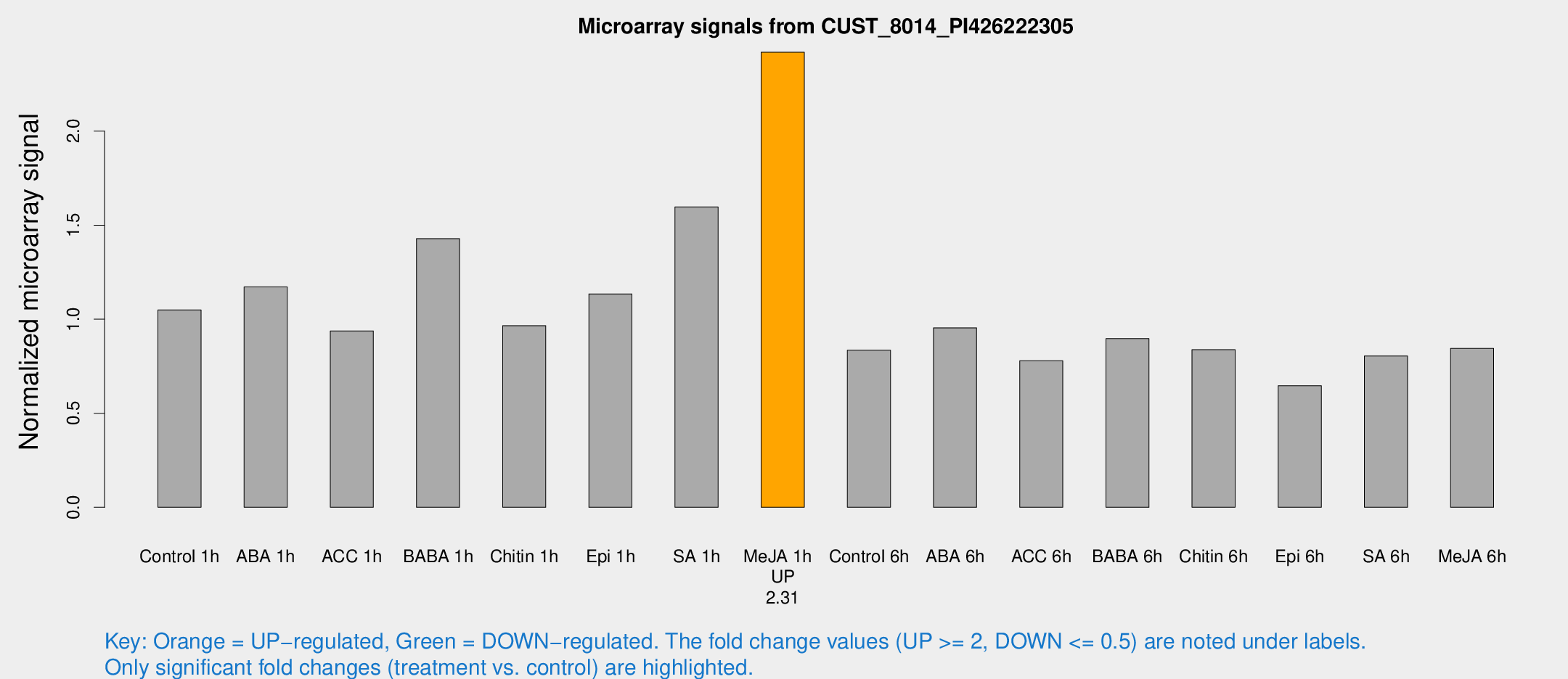
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 377.178 | 41.7721 | 1.04903 | 0.0612546 |
| ABA 1h | 369.783 | 25.2041 | 1.17132 | 0.068346 |
| ACC 1h | 370.498 | 102.061 | 0.937347 | 0.2204 |
| BABA 1h | 508.541 | 93.885 | 1.42868 | 0.146291 |
| Chitin 1h | 312.457 | 33.9685 | 0.965726 | 0.0763224 |
| Epi 1h | 348.989 | 20.3969 | 1.13458 | 0.0662975 |
| SA 1h | 637.937 | 195.791 | 1.59655 | 0.475056 |
| Me-JA 1h | 713.612 | 94.9002 | 2.41941 | 0.14016 |
| Control 6h | 322.457 | 80.5109 | 0.834853 | 0.181021 |
| ABA 6h | 369.556 | 57.7301 | 0.954207 | 0.0997262 |
| ACC 6h | 318.984 | 21.7848 | 0.778891 | 0.105494 |
| BABA 6h | 364.874 | 53.0399 | 0.896789 | 0.102282 |
| Chitin 6h | 319.492 | 28.5968 | 0.837746 | 0.0517945 |
| Epi 6h | 269.211 | 49.7517 | 0.64668 | 0.0983975 |
| SA 6h | 296.024 | 61.5223 | 0.804911 | 0.0956968 |
| Me-JA 6h | 315.611 | 66.8862 | 0.844511 | 0.141978 |
Source Transcript PGSC0003DMT400039875 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G13010.1 | +1 | 0.0 | 1263 | 645/811 (80%) | RNA helicase family protein | chr5:4122747-4128660 FORWARD LENGTH=1255 |
