Probe CUST_7983_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7983_PI426222305 | JHI_St_60k_v1 | DMT400039835 | GCCAAAATATCGATAACCTTCTTGCTCATTTGTAATCACAATTCTCCCCTGCACTTTTTC |
All Microarray Probes Designed to Gene DMG400015403
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7983_PI426222305 | JHI_St_60k_v1 | DMT400039835 | GCCAAAATATCGATAACCTTCTTGCTCATTTGTAATCACAATTCTCCCCTGCACTTTTTC |
Microarray Signals from CUST_7983_PI426222305
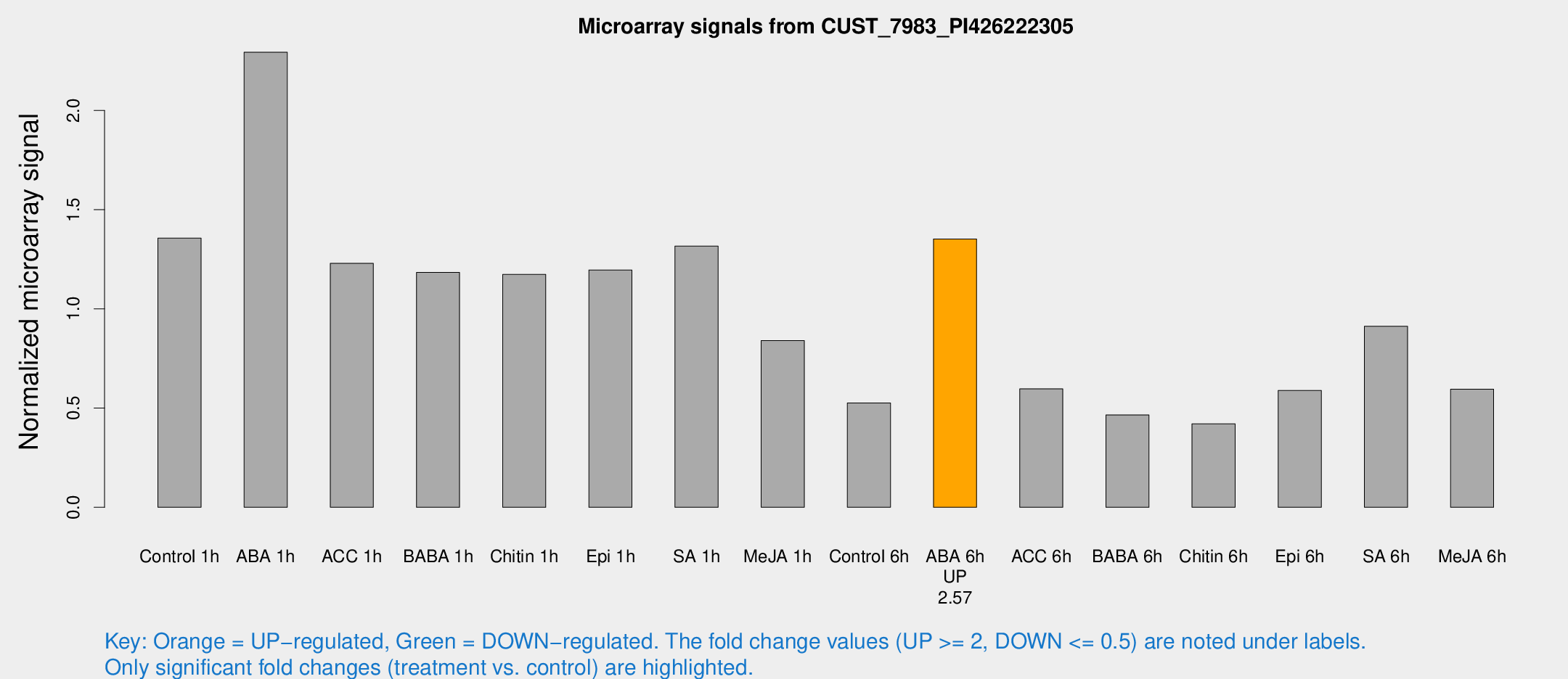
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 280.617 | 16.5063 | 1.35656 | 0.0796858 |
| ABA 1h | 422.06 | 34.9578 | 2.29335 | 0.133392 |
| ACC 1h | 264.36 | 31.1339 | 1.22911 | 0.0729529 |
| BABA 1h | 244.41 | 43.4453 | 1.18377 | 0.111323 |
| Chitin 1h | 221.222 | 28.0743 | 1.17335 | 0.0752998 |
| Epi 1h | 215.305 | 18.6376 | 1.19573 | 0.122766 |
| SA 1h | 286.242 | 44.8107 | 1.31593 | 0.127265 |
| Me-JA 1h | 142.559 | 10.9261 | 0.84048 | 0.0518702 |
| Control 6h | 117.825 | 30.6664 | 0.525709 | 0.114731 |
| ABA 6h | 299.649 | 27.4435 | 1.3524 | 0.0794893 |
| ACC 6h | 142.397 | 11.5418 | 0.596822 | 0.074437 |
| BABA 6h | 109.363 | 13.9355 | 0.465225 | 0.04916 |
| Chitin 6h | 94.374 | 13.3515 | 0.420687 | 0.0557647 |
| Epi 6h | 137.649 | 9.23633 | 0.588427 | 0.0393241 |
| SA 6h | 190.906 | 29.1381 | 0.912113 | 0.253917 |
| Me-JA 6h | 123.823 | 13.572 | 0.595254 | 0.040836 |
Source Transcript PGSC0003DMT400039835 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G37360.1 | +3 | 9e-151 | 445 | 230/469 (49%) | cytochrome P450, family 81, subfamily D, polypeptide 2 | chr4:17567124-17568858 REVERSE LENGTH=499 |
