Probe CUST_7791_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7791_PI426222305 | JHI_St_60k_v1 | DMT400013206 | GACATGTGTTGTTAGTGCCTCCTCTCAAGCACACATCTATAAAGAAGATGGCCTGGTCAT |
All Microarray Probes Designed to Gene DMG400005157
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7522_PI426222305 | JHI_St_60k_v1 | DMT400013207 | TTAACGTCTTTACTAGTTTGAATCTAGAATTGTGTAATTTGCGCCCCTACTTCTCTGATT |
| CUST_7702_PI426222305 | JHI_St_60k_v1 | DMT400013208 | AGTACACTGTATTTCATATCCCCCTTCACTGTATTTTGTGTCAAATCTACATTTCAGAAG |
| CUST_7791_PI426222305 | JHI_St_60k_v1 | DMT400013206 | GACATGTGTTGTTAGTGCCTCCTCTCAAGCACACATCTATAAAGAAGATGGCCTGGTCAT |
Microarray Signals from CUST_7791_PI426222305
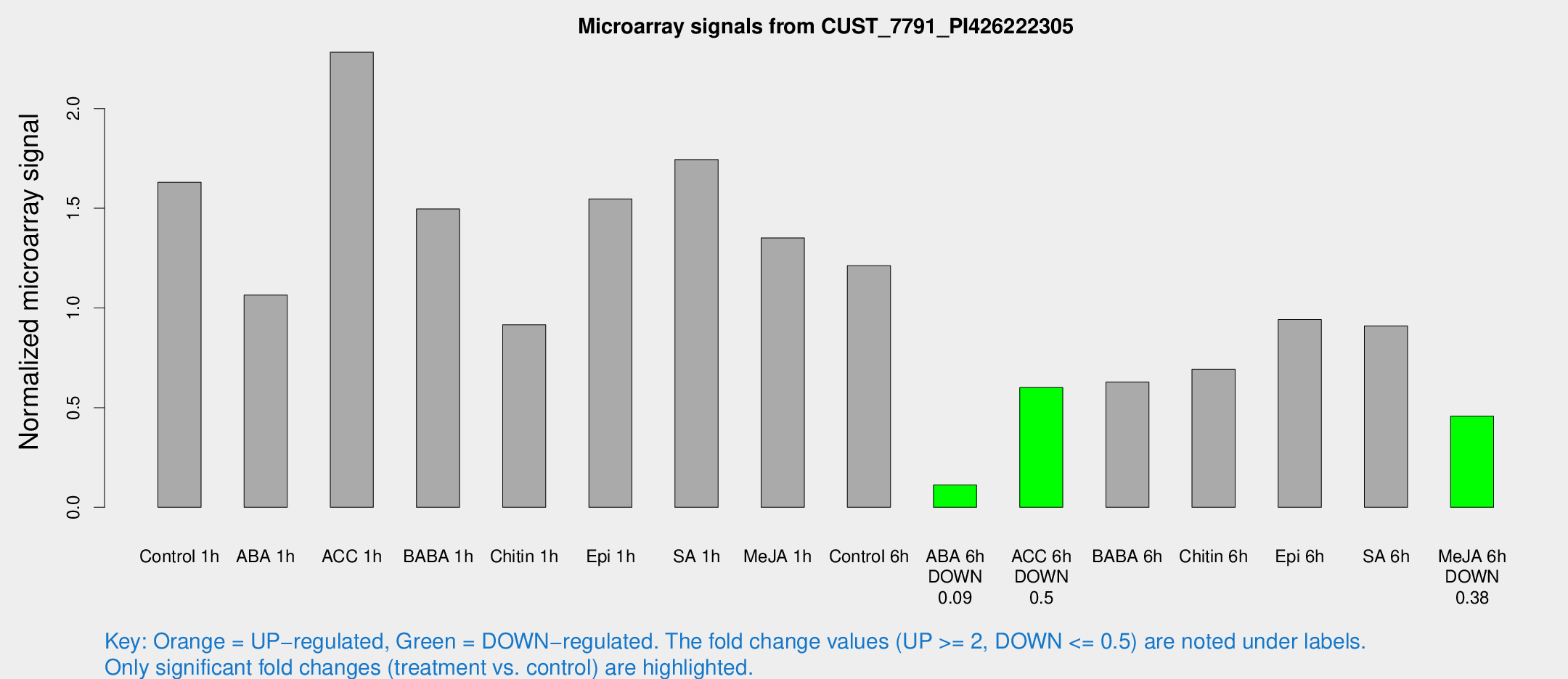
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 92.2492 | 27.9003 | 1.63013 | 0.471735 |
| ABA 1h | 51.203 | 13.0668 | 1.0647 | 0.263862 |
| ACC 1h | 127.932 | 30.0549 | 2.28271 | 0.469752 |
| BABA 1h | 79.6441 | 19.8578 | 1.49653 | 0.304517 |
| Chitin 1h | 44.3607 | 11.1436 | 0.915494 | 0.24438 |
| Epi 1h | 69.8899 | 11.6795 | 1.54656 | 0.280077 |
| SA 1h | 94.0015 | 16.9919 | 1.74458 | 0.342583 |
| Me-JA 1h | 58.7903 | 12.4898 | 1.35102 | 0.264415 |
| Control 6h | 64.1895 | 13.1249 | 1.21156 | 0.219447 |
| ABA 6h | 6.07509 | 3.5313 | 0.112206 | 0.065166 |
| ACC 6h | 35.5769 | 4.52804 | 0.60072 | 0.105532 |
| BABA 6h | 37.4263 | 7.31201 | 0.627644 | 0.118811 |
| Chitin 6h | 41.7368 | 13.9468 | 0.691871 | 0.225507 |
| Epi 6h | 54.0924 | 5.06845 | 0.941733 | 0.0886003 |
| SA 6h | 48.0411 | 10.2246 | 0.909976 | 0.246094 |
| Me-JA 6h | 24.0089 | 4.67071 | 0.457019 | 0.079209 |
Source Transcript PGSC0003DMT400013206 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
