Probe CUST_7702_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7702_PI426222305 | JHI_St_60k_v1 | DMT400013208 | AGTACACTGTATTTCATATCCCCCTTCACTGTATTTTGTGTCAAATCTACATTTCAGAAG |
All Microarray Probes Designed to Gene DMG400005157
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7791_PI426222305 | JHI_St_60k_v1 | DMT400013206 | GACATGTGTTGTTAGTGCCTCCTCTCAAGCACACATCTATAAAGAAGATGGCCTGGTCAT |
| CUST_7522_PI426222305 | JHI_St_60k_v1 | DMT400013207 | TTAACGTCTTTACTAGTTTGAATCTAGAATTGTGTAATTTGCGCCCCTACTTCTCTGATT |
| CUST_7702_PI426222305 | JHI_St_60k_v1 | DMT400013208 | AGTACACTGTATTTCATATCCCCCTTCACTGTATTTTGTGTCAAATCTACATTTCAGAAG |
Microarray Signals from CUST_7702_PI426222305
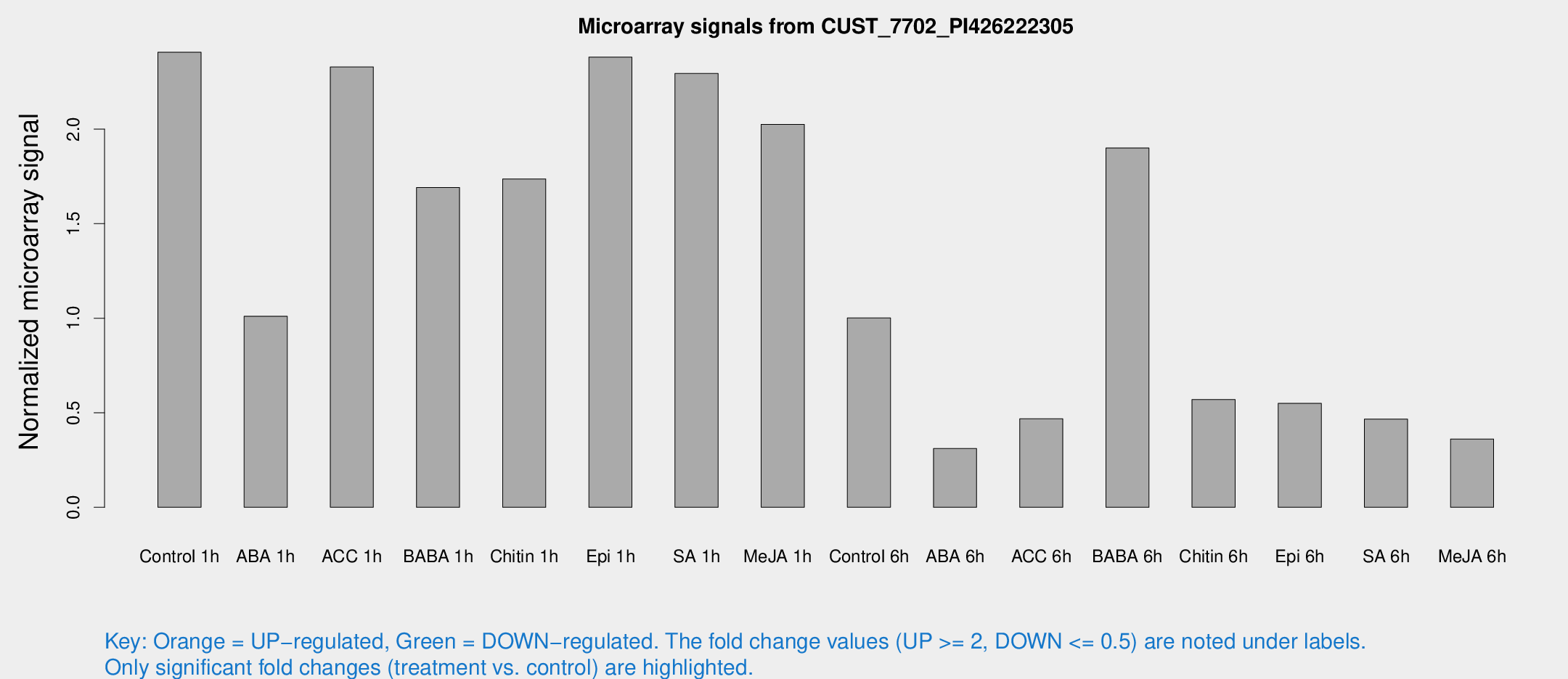
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 38.7858 | 3.79482 | 2.4066 | 0.23709 |
| ABA 1h | 15.8501 | 5.40876 | 1.01034 | 0.350944 |
| ACC 1h | 47.0713 | 17.9976 | 2.32878 | 1.14117 |
| BABA 1h | 28.6383 | 8.37038 | 1.69125 | 0.396435 |
| Chitin 1h | 26.1361 | 5.16199 | 1.73661 | 0.550125 |
| Epi 1h | 33.2904 | 3.61927 | 2.38078 | 0.247752 |
| SA 1h | 43.4646 | 14.2021 | 2.29398 | 0.923044 |
| Me-JA 1h | 28.9993 | 8.04834 | 2.02533 | 0.671703 |
| Control 6h | 16.7971 | 3.79469 | 1.0012 | 0.204928 |
| ABA 6h | 5.24147 | 3.04364 | 0.310744 | 0.178862 |
| ACC 6h | 9.43008 | 3.57981 | 0.468339 | 0.211733 |
| BABA 6h | 75.7307 | 58.5595 | 1.90005 | 4.59352 |
| Chitin 6h | 10.7886 | 3.46571 | 0.569746 | 0.219893 |
| Epi 6h | 10.1773 | 3.4291 | 0.549651 | 0.188914 |
| SA 6h | 7.8378 | 3.13366 | 0.466752 | 0.214036 |
| Me-JA 6h | 5.8183 | 2.91957 | 0.361207 | 0.187003 |
Source Transcript PGSC0003DMT400013208 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
