Probe CUST_7602_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7602_PI426222305 | JHI_St_60k_v1 | DMT400025820 | GTATCAATGGCATATGCTAAACCATCTTGTTCATGGATTGGATTAGGAAAGAAGACAAAA |
All Microarray Probes Designed to Gene DMG400009973
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7602_PI426222305 | JHI_St_60k_v1 | DMT400025820 | GTATCAATGGCATATGCTAAACCATCTTGTTCATGGATTGGATTAGGAAAGAAGACAAAA |
Microarray Signals from CUST_7602_PI426222305
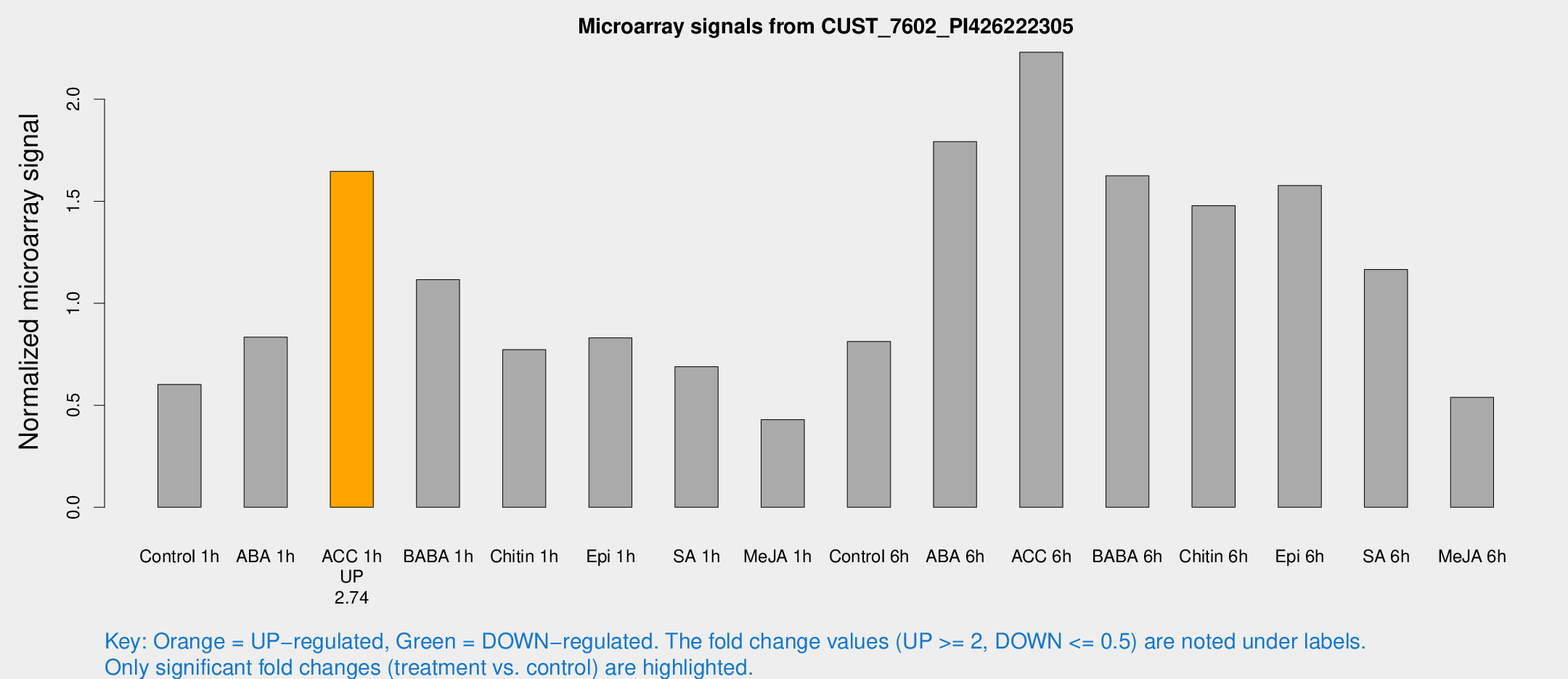
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 656.667 | 122.443 | 0.601706 | 0.0969111 |
| ABA 1h | 794.122 | 107.565 | 0.834303 | 0.0858066 |
| ACC 1h | 1889.39 | 432.301 | 1.64627 | 0.288622 |
| BABA 1h | 1136.54 | 65.8834 | 1.11541 | 0.0747178 |
| Chitin 1h | 762.664 | 159.729 | 0.772522 | 0.181866 |
| Epi 1h | 759.661 | 44.0748 | 0.83063 | 0.0515362 |
| SA 1h | 749.254 | 45.3339 | 0.689356 | 0.0399019 |
| Me-JA 1h | 373.476 | 34.9946 | 0.429883 | 0.0251002 |
| Control 6h | 1078.52 | 398.477 | 0.812723 | 0.416106 |
| ABA 6h | 2169.04 | 603.352 | 1.79219 | 0.597628 |
| ACC 6h | 2886.75 | 721.226 | 2.23029 | 0.356366 |
| BABA 6h | 1974.98 | 354.815 | 1.62465 | 0.234423 |
| Chitin 6h | 1712.08 | 288.051 | 1.47798 | 0.3183 |
| Epi 6h | 2003.61 | 538.627 | 1.57663 | 0.452679 |
| SA 6h | 1252.2 | 200.789 | 1.16569 | 0.320422 |
| Me-JA 6h | 624.673 | 169.544 | 0.538824 | 0.15067 |
Source Transcript PGSC0003DMT400025820 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G76240.1 | +2 | 1e-78 | 248 | 152/311 (49%) | Arabidopsis protein of unknown function (DUF241) | chr1:28602949-28603875 REVERSE LENGTH=308 |
