Probe CUST_6725_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6725_PI426222305 | JHI_St_60k_v1 | DMT400014621 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
All Microarray Probes Designed to Gene DMG400005718
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6456_PI426222305 | JHI_St_60k_v1 | DMT400014625 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6725_PI426222305 | JHI_St_60k_v1 | DMT400014621 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6802_PI426222305 | JHI_St_60k_v1 | DMT400014622 | TTCCTAGTGAGTCTTACCTGCTTATTACTTCGTTCATGTAAAACAGGTTTCAGTTTATAG |
| CUST_6521_PI426222305 | JHI_St_60k_v1 | DMT400014626 | CGCAAAGAGTTGATTTTACTCTAATGCTCAACTGGTATCTGTTTTGATCTGTAGTTCTTT |
| CUST_6311_PI426222305 | JHI_St_60k_v1 | DMT400014623 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6733_PI426222305 | JHI_St_60k_v1 | DMT400014624 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6666_PI426222305 | JHI_St_60k_v1 | DMT400014627 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
Microarray Signals from CUST_6725_PI426222305
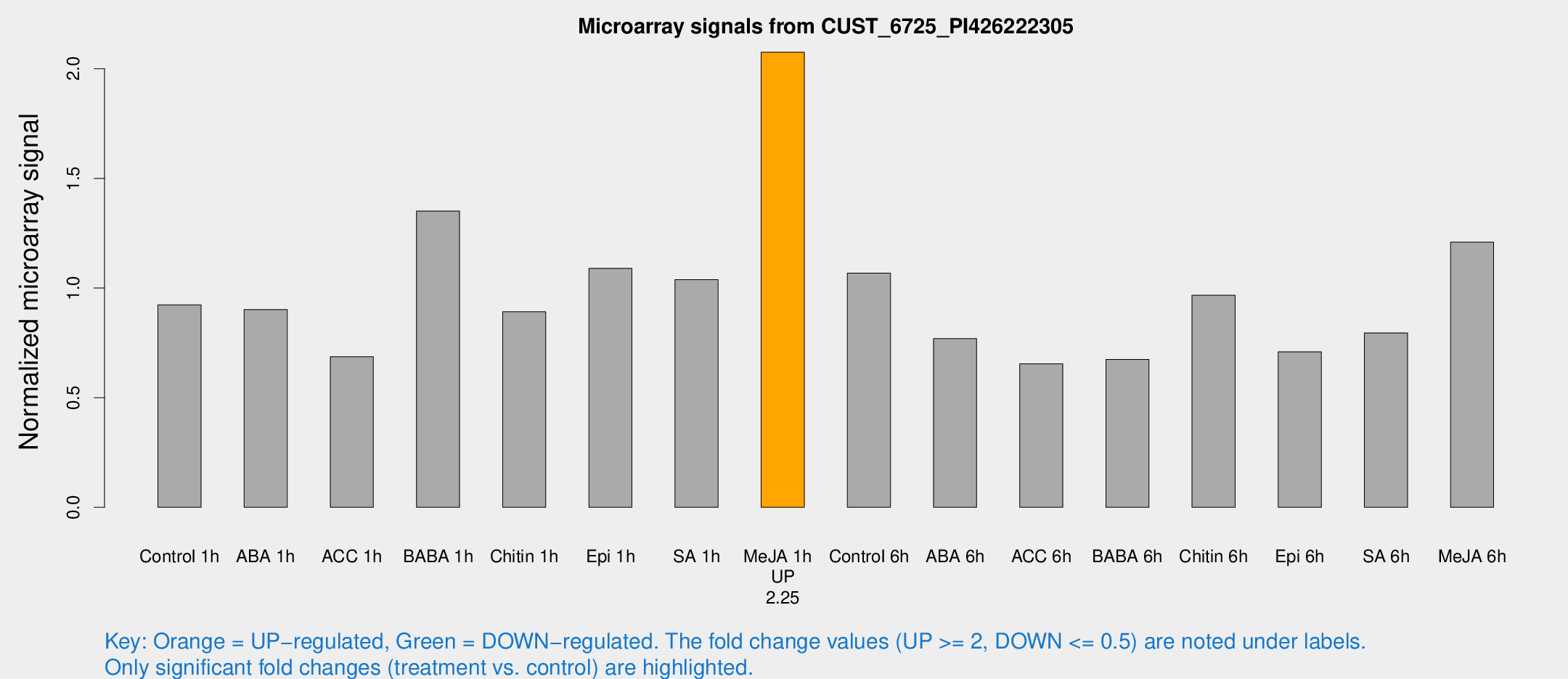
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 51.5993 | 7.62266 | 0.923422 | 0.0817607 |
| ABA 1h | 44.7091 | 7.09101 | 0.901539 | 0.0858781 |
| ACC 1h | 42.6815 | 13.6653 | 0.687044 | 0.190795 |
| BABA 1h | 71.7069 | 5.54018 | 1.35126 | 0.105083 |
| Chitin 1h | 44.9194 | 7.17873 | 0.891497 | 0.0846784 |
| Epi 1h | 52.1294 | 5.16161 | 1.08966 | 0.107289 |
| SA 1h | 59.868 | 9.14062 | 1.03806 | 0.170004 |
| Me-JA 1h | 94.46 | 12.8962 | 2.0754 | 0.142558 |
| Control 6h | 62.9917 | 15.9432 | 1.06775 | 0.219444 |
| ABA 6h | 47.3471 | 10.0939 | 0.769139 | 0.162559 |
| ACC 6h | 43.4977 | 9.90786 | 0.65421 | 0.166495 |
| BABA 6h | 47.2224 | 14.3192 | 0.674735 | 0.26794 |
| Chitin 6h | 56.6007 | 5.02654 | 0.966768 | 0.0997886 |
| Epi 6h | 50.66 | 18.7741 | 0.708921 | 0.347455 |
| SA 6h | 53.5471 | 23.1103 | 0.795108 | 0.627669 |
| Me-JA 6h | 69.5087 | 14.772 | 1.20913 | 0.193823 |
Source Transcript PGSC0003DMT400014621 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73875.1 | +2 | 9e-114 | 352 | 180/325 (55%) | DNAse I-like superfamily protein | chr1:27781112-27784571 FORWARD LENGTH=454 |
