Probe CUST_6666_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6666_PI426222305 | JHI_St_60k_v1 | DMT400014627 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
All Microarray Probes Designed to Gene DMG400005718
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6802_PI426222305 | JHI_St_60k_v1 | DMT400014622 | TTCCTAGTGAGTCTTACCTGCTTATTACTTCGTTCATGTAAAACAGGTTTCAGTTTATAG |
| CUST_6666_PI426222305 | JHI_St_60k_v1 | DMT400014627 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6456_PI426222305 | JHI_St_60k_v1 | DMT400014625 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6733_PI426222305 | JHI_St_60k_v1 | DMT400014624 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6311_PI426222305 | JHI_St_60k_v1 | DMT400014623 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6725_PI426222305 | JHI_St_60k_v1 | DMT400014621 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6521_PI426222305 | JHI_St_60k_v1 | DMT400014626 | CGCAAAGAGTTGATTTTACTCTAATGCTCAACTGGTATCTGTTTTGATCTGTAGTTCTTT |
Microarray Signals from CUST_6666_PI426222305
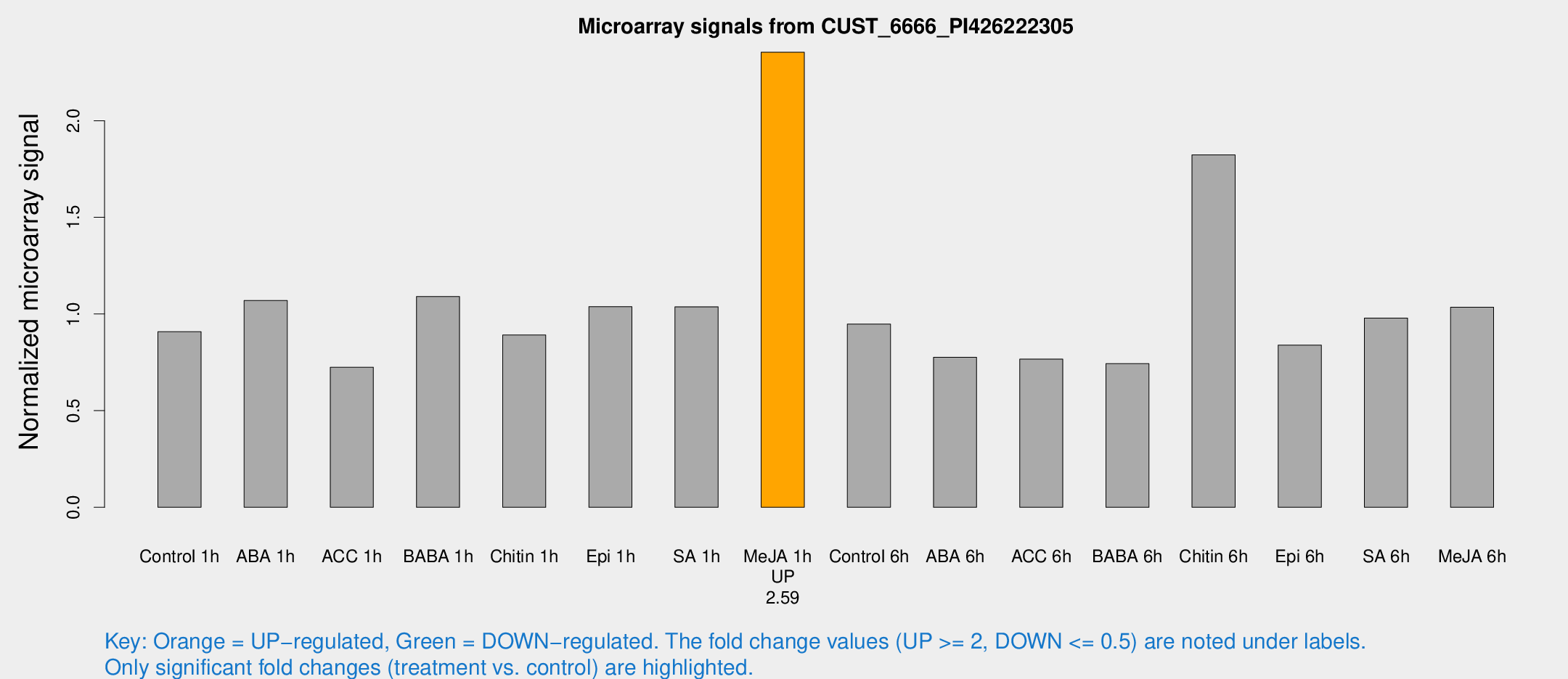
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 52.0179 | 11.1768 | 0.908141 | 0.131582 |
| ABA 1h | 55.2874 | 12.9984 | 1.07014 | 0.205481 |
| ACC 1h | 49.0081 | 17.3821 | 0.72455 | 0.318104 |
| BABA 1h | 59.1208 | 9.02929 | 1.09026 | 0.0991404 |
| Chitin 1h | 44.2441 | 4.38433 | 0.891226 | 0.0875562 |
| Epi 1h | 49.4372 | 4.62757 | 1.03811 | 0.0978785 |
| SA 1h | 59.2006 | 6.12707 | 1.03744 | 0.0897555 |
| Me-JA 1h | 105.97 | 7.09572 | 2.35421 | 0.156601 |
| Control 6h | 56.2613 | 13.8306 | 0.947564 | 0.195953 |
| ABA 6h | 46.7415 | 7.81771 | 0.775782 | 0.11768 |
| ACC 6h | 50.2679 | 9.25239 | 0.766623 | 0.138277 |
| BABA 6h | 50.8037 | 14.7833 | 0.743109 | 0.242324 |
| Chitin 6h | 182.924 | 130.026 | 1.82302 | 1.88824 |
| Epi 6h | 55.269 | 13.7056 | 0.838979 | 0.254799 |
| SA 6h | 66.9829 | 28.9695 | 0.979157 | 0.810546 |
| Me-JA 6h | 61.9261 | 17.3893 | 1.03467 | 0.242037 |
Source Transcript PGSC0003DMT400014627 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73875.1 | +2 | 5e-131 | 395 | 202/352 (57%) | DNAse I-like superfamily protein | chr1:27781112-27784571 FORWARD LENGTH=454 |
