Probe CUST_6521_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6521_PI426222305 | JHI_St_60k_v1 | DMT400014626 | CGCAAAGAGTTGATTTTACTCTAATGCTCAACTGGTATCTGTTTTGATCTGTAGTTCTTT |
All Microarray Probes Designed to Gene DMG400005718
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6802_PI426222305 | JHI_St_60k_v1 | DMT400014622 | TTCCTAGTGAGTCTTACCTGCTTATTACTTCGTTCATGTAAAACAGGTTTCAGTTTATAG |
| CUST_6725_PI426222305 | JHI_St_60k_v1 | DMT400014621 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6311_PI426222305 | JHI_St_60k_v1 | DMT400014623 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6456_PI426222305 | JHI_St_60k_v1 | DMT400014625 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6521_PI426222305 | JHI_St_60k_v1 | DMT400014626 | CGCAAAGAGTTGATTTTACTCTAATGCTCAACTGGTATCTGTTTTGATCTGTAGTTCTTT |
| CUST_6666_PI426222305 | JHI_St_60k_v1 | DMT400014627 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6733_PI426222305 | JHI_St_60k_v1 | DMT400014624 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
Microarray Signals from CUST_6521_PI426222305
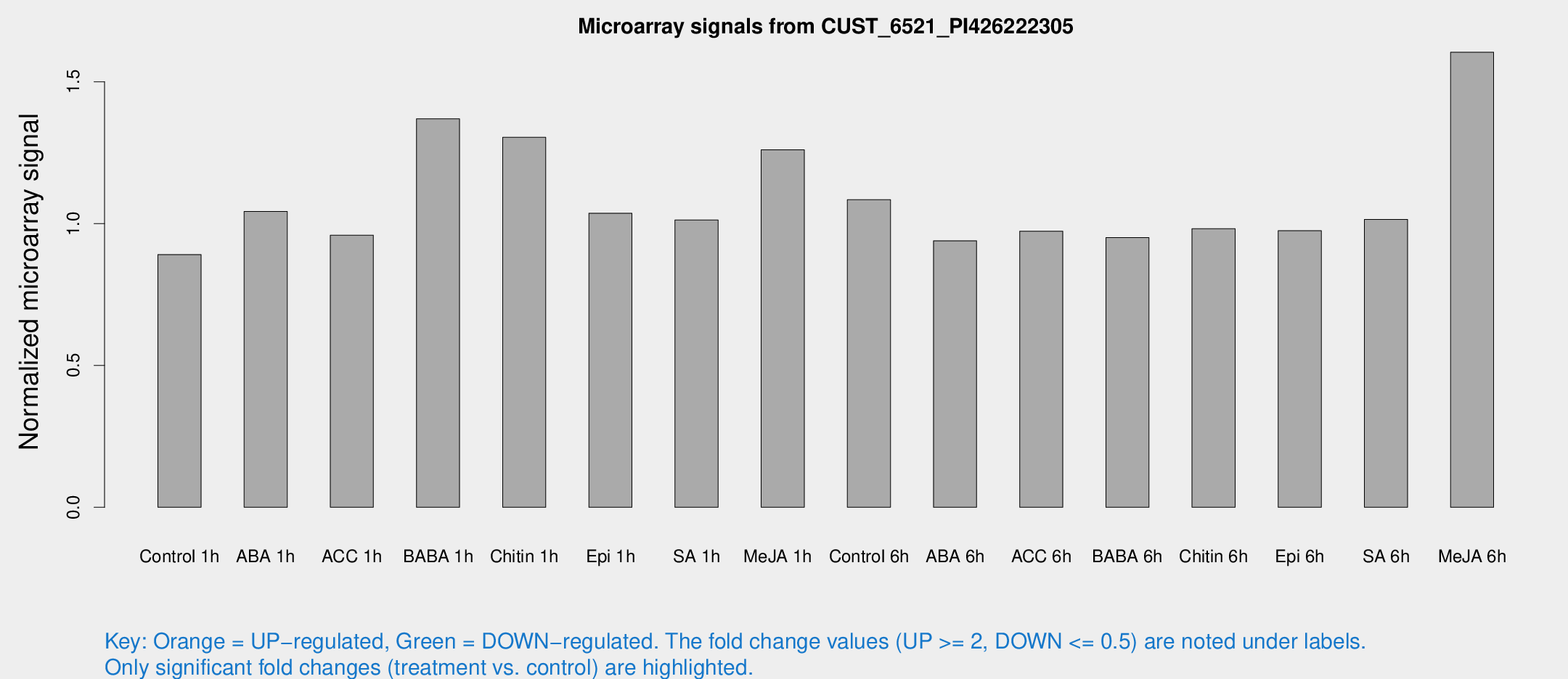
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.18437 | 3.58161 | 0.89101 | 0.516012 |
| ABA 1h | 6.4061 | 3.51565 | 1.04255 | 0.57351 |
| ACC 1h | 6.85439 | 3.98115 | 0.959122 | 0.555676 |
| BABA 1h | 9.48896 | 3.93233 | 1.36924 | 0.608559 |
| Chitin 1h | 8.476 | 3.7254 | 1.3041 | 0.63392 |
| Epi 1h | 6.23669 | 3.62357 | 1.0366 | 0.601197 |
| SA 1h | 7.48442 | 3.60479 | 1.01255 | 0.517179 |
| Me-JA 1h | 7.39101 | 3.67813 | 1.26046 | 0.655654 |
| Control 6h | 7.62669 | 3.89892 | 1.08416 | 0.568083 |
| ABA 6h | 6.9424 | 4.03008 | 0.939464 | 0.544583 |
| ACC 6h | 7.94155 | 4.71767 | 0.973476 | 0.563711 |
| BABA 6h | 7.42779 | 4.26435 | 0.950669 | 0.544433 |
| Chitin 6h | 7.25651 | 4.20426 | 0.982044 | 0.568928 |
| Epi 6h | 7.70049 | 4.50211 | 0.974793 | 0.564784 |
| SA 6h | 6.96763 | 4.03501 | 1.01481 | 0.58766 |
| Me-JA 6h | 13.2392 | 5.71011 | 1.60413 | 0.699786 |
Source Transcript PGSC0003DMT400014626 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73875.1 | +1 | 1e-82 | 266 | 132/213 (62%) | DNAse I-like superfamily protein | chr1:27781112-27784571 FORWARD LENGTH=454 |
