Probe CUST_6492_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6492_PI426222305 | JHI_St_60k_v1 | DMT400014620 | ACTGACAACGAATTTGCAATCGCTTTTTTGGAGAATTATAGCGGCTACTATTTTGTTAGG |
All Microarray Probes Designed to Gene DMG400005717
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6492_PI426222305 | JHI_St_60k_v1 | DMT400014620 | ACTGACAACGAATTTGCAATCGCTTTTTTGGAGAATTATAGCGGCTACTATTTTGTTAGG |
Microarray Signals from CUST_6492_PI426222305
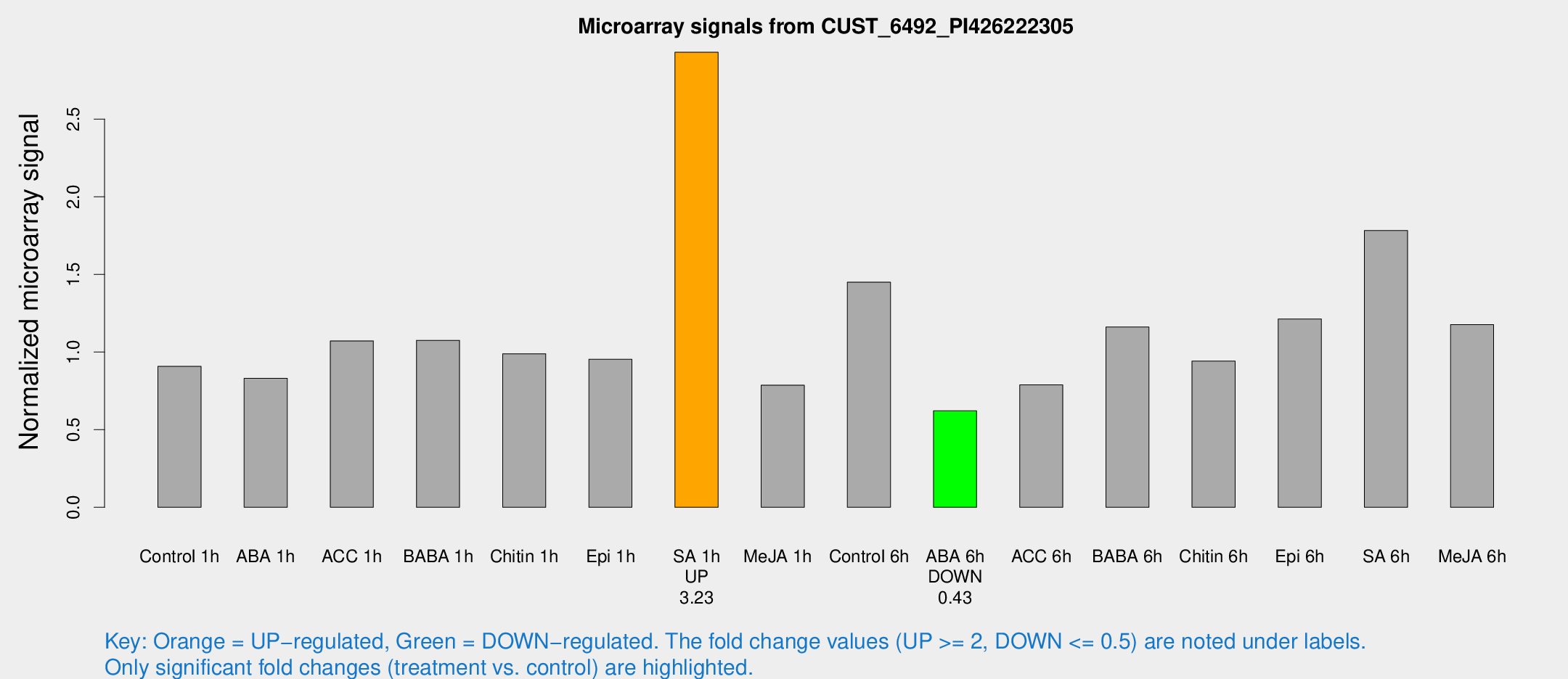
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6537.88 | 952.697 | 0.907784 | 0.0747592 |
| ABA 1h | 5338.32 | 920.527 | 0.830228 | 0.135291 |
| ACC 1h | 8233.48 | 2004.4 | 1.07135 | 0.208731 |
| BABA 1h | 7732.81 | 1722.28 | 1.07522 | 0.158276 |
| Chitin 1h | 6350.72 | 695.09 | 0.988775 | 0.0570897 |
| Epi 1h | 5828.08 | 336.672 | 0.953141 | 0.0550327 |
| SA 1h | 21866.6 | 3517.24 | 2.93045 | 0.329249 |
| Me-JA 1h | 4646.27 | 699.436 | 0.786643 | 0.0592051 |
| Control 6h | 10760.3 | 2126.54 | 1.45009 | 0.196119 |
| ABA 6h | 4734.47 | 600.196 | 0.621108 | 0.0543633 |
| ACC 6h | 6411.13 | 395.915 | 0.788476 | 0.11757 |
| BABA 6h | 9175.82 | 529.803 | 1.16084 | 0.067023 |
| Chitin 6h | 7091.07 | 410.294 | 0.941397 | 0.0543545 |
| Epi 6h | 9679.45 | 560.499 | 1.21209 | 0.0699826 |
| SA 6h | 12499.2 | 795.411 | 1.78248 | 0.153866 |
| Me-JA 6h | 8610.7 | 1787.97 | 1.1769 | 0.175651 |
Source Transcript PGSC0003DMT400014620 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
