Probe CUST_6311_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6311_PI426222305 | JHI_St_60k_v1 | DMT400014623 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
All Microarray Probes Designed to Gene DMG400005718
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6802_PI426222305 | JHI_St_60k_v1 | DMT400014622 | TTCCTAGTGAGTCTTACCTGCTTATTACTTCGTTCATGTAAAACAGGTTTCAGTTTATAG |
| CUST_6521_PI426222305 | JHI_St_60k_v1 | DMT400014626 | CGCAAAGAGTTGATTTTACTCTAATGCTCAACTGGTATCTGTTTTGATCTGTAGTTCTTT |
| CUST_6725_PI426222305 | JHI_St_60k_v1 | DMT400014621 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6456_PI426222305 | JHI_St_60k_v1 | DMT400014625 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6666_PI426222305 | JHI_St_60k_v1 | DMT400014627 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6311_PI426222305 | JHI_St_60k_v1 | DMT400014623 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
| CUST_6733_PI426222305 | JHI_St_60k_v1 | DMT400014624 | CAATTGCAGATGATAAGCGCAAGTGGGTTTACTCTACTCACGTCGTTTCCCCAAATCAAG |
Microarray Signals from CUST_6311_PI426222305
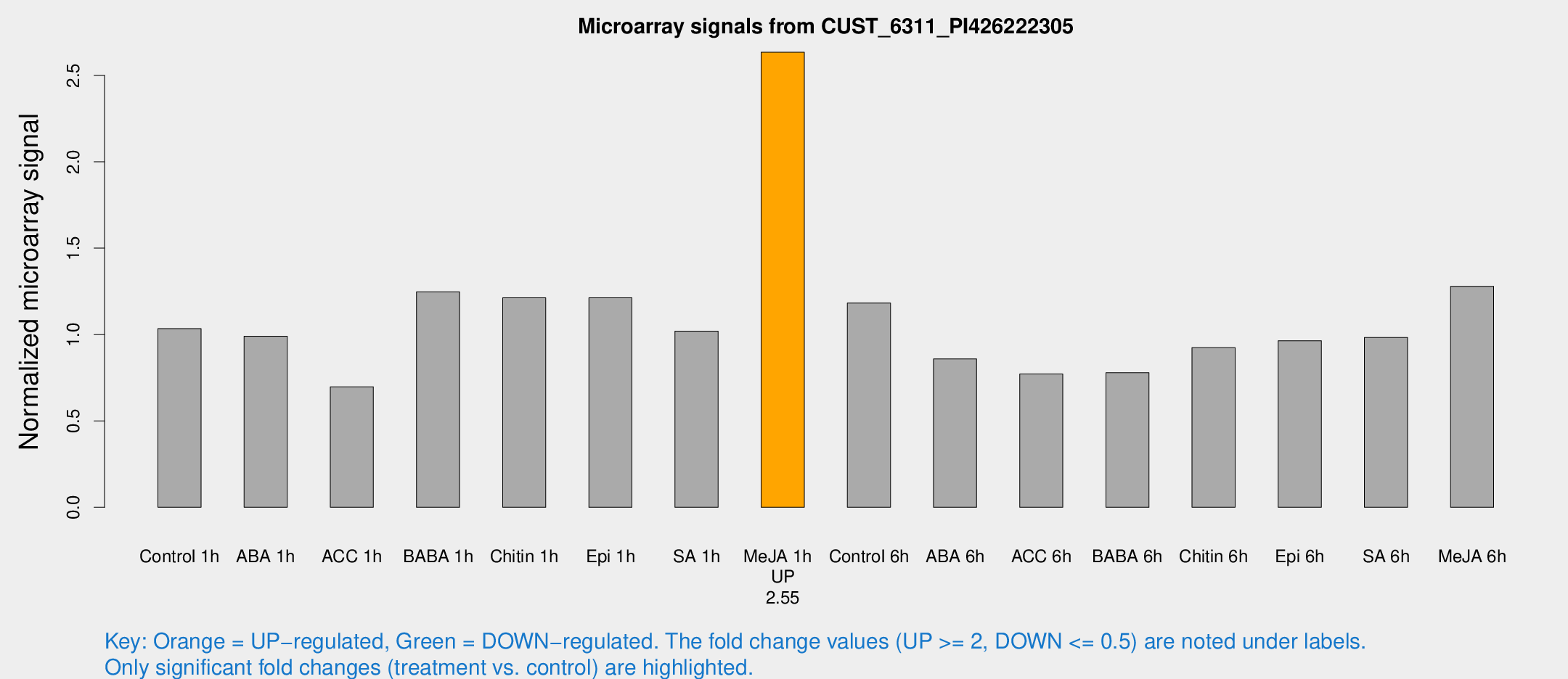
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 52.7021 | 7.17453 | 1.03425 | 0.0886831 |
| ABA 1h | 46.3489 | 10.1042 | 0.990016 | 0.181044 |
| ACC 1h | 45.8302 | 19.5124 | 0.697378 | 0.383885 |
| BABA 1h | 63.9511 | 14.6026 | 1.24744 | 0.195688 |
| Chitin 1h | 56.3003 | 10.2271 | 1.21284 | 0.137068 |
| Epi 1h | 56.6628 | 14.3919 | 1.21262 | 0.354719 |
| SA 1h | 52.5014 | 4.35998 | 1.01947 | 0.0846958 |
| Me-JA 1h | 109.078 | 12.6901 | 2.6342 | 0.171247 |
| Control 6h | 62.8861 | 14.5327 | 1.18201 | 0.207921 |
| ABA 6h | 47.2051 | 8.14734 | 0.85873 | 0.111092 |
| ACC 6h | 46.2665 | 9.05599 | 0.771923 | 0.0936979 |
| BABA 6h | 45.0654 | 7.75209 | 0.778944 | 0.110569 |
| Chitin 6h | 49.277 | 4.57517 | 0.924102 | 0.0858925 |
| Epi 6h | 57.1893 | 12.4161 | 0.964263 | 0.183695 |
| SA 6h | 53.6149 | 16.1832 | 0.983203 | 0.479291 |
| Me-JA 6h | 67.4658 | 14.3638 | 1.27906 | 0.223583 |
Source Transcript PGSC0003DMT400014623 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73875.1 | +2 | 7e-85 | 276 | 136/233 (58%) | DNAse I-like superfamily protein | chr1:27781112-27784571 FORWARD LENGTH=454 |
