Probe CUST_6048_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6048_PI426222305 | JHI_St_60k_v1 | DMT400010699 | TGTTTTCCTTTTGTAGAGTGCAACGAGTGTACCAACAACTTCGACTTGCCCTCAGCTCTA |
All Microarray Probes Designed to Gene DMG401004173
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6048_PI426222305 | JHI_St_60k_v1 | DMT400010699 | TGTTTTCCTTTTGTAGAGTGCAACGAGTGTACCAACAACTTCGACTTGCCCTCAGCTCTA |
| CUST_6065_PI426222305 | JHI_St_60k_v1 | DMT400010693 | GATCCAACAATATCGTGTCGGAACGTGAATGGGACACTTTGTGAGAGATTACCCTAGAAC |
Microarray Signals from CUST_6048_PI426222305
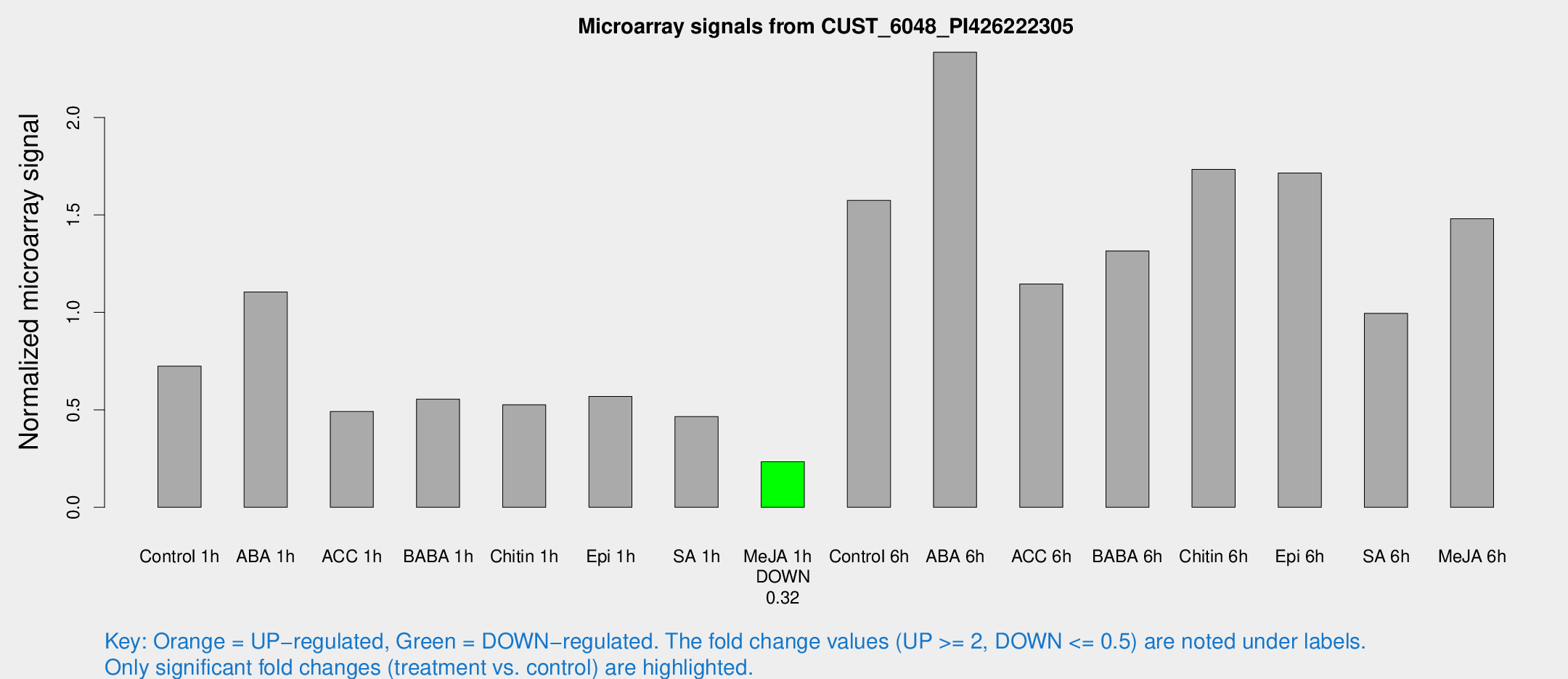
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 194.419 | 13.2158 | 0.723964 | 0.043856 |
| ABA 1h | 261.42 | 15.4804 | 1.10495 | 0.0653853 |
| ACC 1h | 136.52 | 15.4623 | 0.491299 | 0.0335108 |
| BABA 1h | 147.851 | 26.6877 | 0.555098 | 0.0572716 |
| Chitin 1h | 126.868 | 8.15112 | 0.525793 | 0.033652 |
| Epi 1h | 133.389 | 14.9593 | 0.568866 | 0.0575145 |
| SA 1h | 130.671 | 18.5598 | 0.465256 | 0.0508051 |
| Me-JA 1h | 51.4937 | 4.93575 | 0.233764 | 0.0252609 |
| Control 6h | 436.563 | 76.768 | 1.57492 | 0.175193 |
| ABA 6h | 688.219 | 122.697 | 2.33463 | 0.354917 |
| ACC 6h | 355.435 | 37.4791 | 1.14538 | 0.157925 |
| BABA 6h | 405.538 | 72.8797 | 1.31513 | 0.212189 |
| Chitin 6h | 493.313 | 28.8252 | 1.73343 | 0.101266 |
| Epi 6h | 529.536 | 86.1417 | 1.71468 | 0.297718 |
| SA 6h | 314.07 | 136.004 | 0.994728 | 0.364122 |
| Me-JA 6h | 420.038 | 106.688 | 1.48026 | 0.256496 |
Source Transcript PGSC0003DMT400010699 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
