Probe CUST_5952_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5952_PI426222305 | JHI_St_60k_v1 | DMT400013055 | TTTGGGTTTGATCTCAACTTTCTGTAGTTCATTTTGTTCAATAAGAAATGTGGATAGCCG |
All Microarray Probes Designed to Gene DMG400005092
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5952_PI426222305 | JHI_St_60k_v1 | DMT400013055 | TTTGGGTTTGATCTCAACTTTCTGTAGTTCATTTTGTTCAATAAGAAATGTGGATAGCCG |
Microarray Signals from CUST_5952_PI426222305
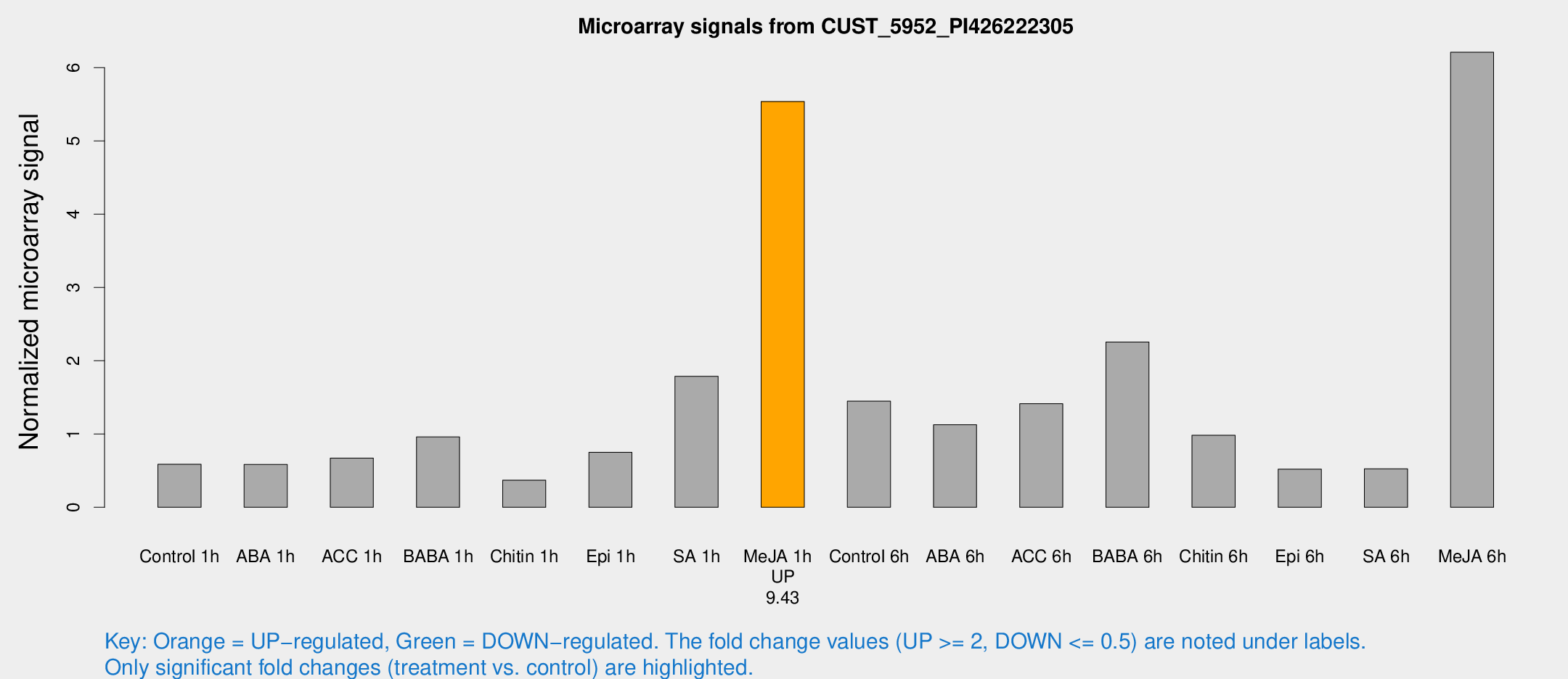
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 17.6936 | 6.63844 | 0.587109 | 0.302685 |
| ABA 1h | 13.3935 | 3.48913 | 0.58558 | 0.166736 |
| ACC 1h | 20.9521 | 8.2236 | 0.67053 | 0.302123 |
| BABA 1h | 25.6436 | 7.60541 | 0.96086 | 0.232698 |
| Chitin 1h | 9.16647 | 3.65362 | 0.370759 | 0.173436 |
| Epi 1h | 17.2566 | 4.14815 | 0.750161 | 0.182077 |
| SA 1h | 48.3109 | 11.1073 | 1.78849 | 0.415756 |
| Me-JA 1h | 114.407 | 10.9644 | 5.53868 | 0.365767 |
| Control 6h | 44.66 | 20.2462 | 1.44808 | 0.596003 |
| ABA 6h | 30.9119 | 4.95482 | 1.12691 | 0.199072 |
| ACC 6h | 41.4555 | 5.90814 | 1.41324 | 0.318268 |
| BABA 6h | 65.7763 | 13.4574 | 2.2567 | 0.371172 |
| Chitin 6h | 26.8615 | 4.41883 | 0.982245 | 0.190848 |
| Epi 6h | 15.7033 | 4.4654 | 0.521057 | 0.178888 |
| SA 6h | 14.1958 | 4.05523 | 0.525857 | 0.215854 |
| Me-JA 6h | 157.101 | 18.7982 | 6.21085 | 0.390033 |
Source Transcript PGSC0003DMT400013055 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G06880.2 | +2 | 1e-61 | 231 | 197/798 (25%) | Transducin/WD40 repeat-like superfamily protein | chr3:2170516-2175686 REVERSE LENGTH=1264 |
