Probe CUST_5662_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5662_PI426222305 | JHI_St_60k_v1 | DMT400062333 | GTGACACTGATTGAAATAGGCCTTTCTTGTGTAGAGCAAGACAGAAACAAAAGGCCTACA |
All Microarray Probes Designed to Gene DMG400024258
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5662_PI426222305 | JHI_St_60k_v1 | DMT400062333 | GTGACACTGATTGAAATAGGCCTTTCTTGTGTAGAGCAAGACAGAAACAAAAGGCCTACA |
Microarray Signals from CUST_5662_PI426222305
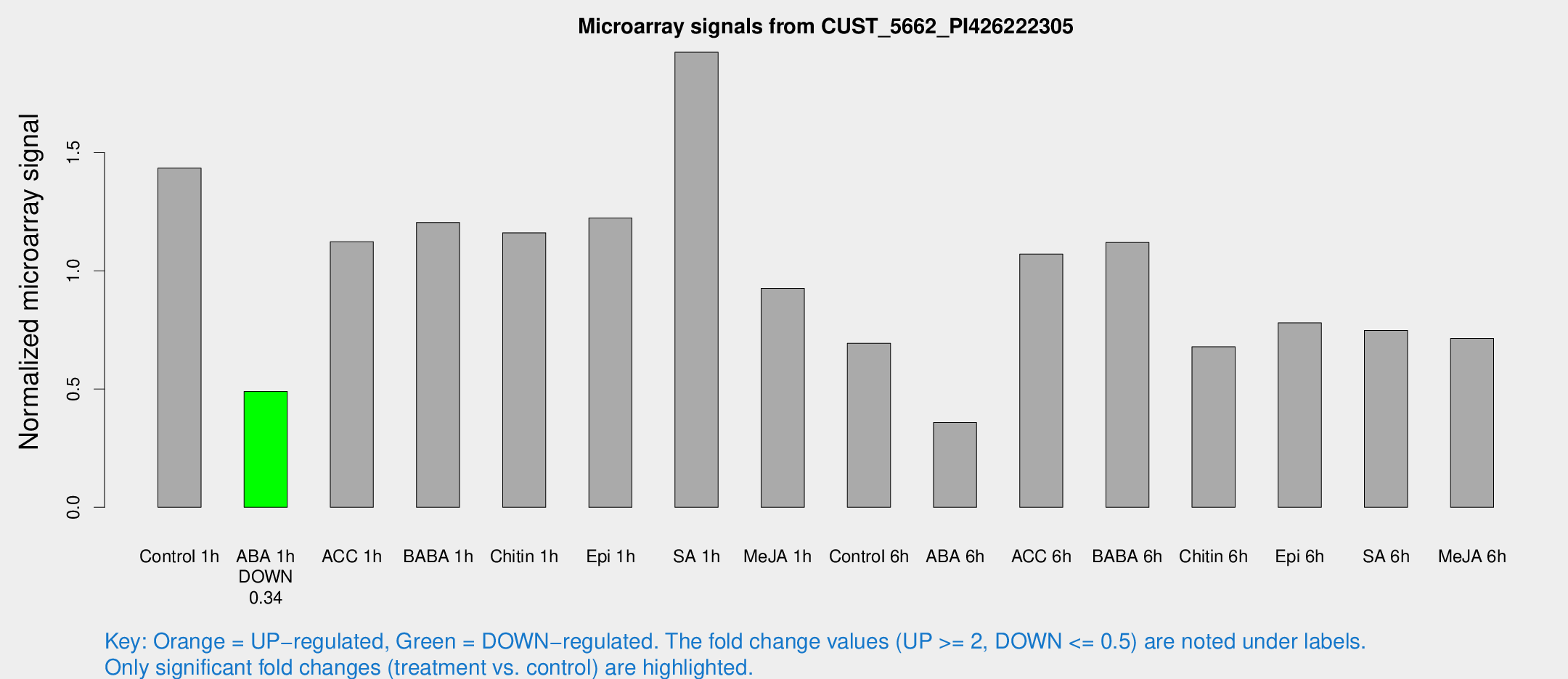
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 382.217 | 70.6196 | 1.43463 | 0.168345 |
| ABA 1h | 115.213 | 18.8785 | 0.490637 | 0.0731897 |
| ACC 1h | 345.02 | 108.299 | 1.12295 | 0.418409 |
| BABA 1h | 315.767 | 66.8656 | 1.20457 | 0.164562 |
| Chitin 1h | 271.485 | 22.4405 | 1.16103 | 0.0684892 |
| Epi 1h | 275.778 | 24.1647 | 1.224 | 0.103993 |
| SA 1h | 518.244 | 60.7897 | 1.92475 | 0.160448 |
| Me-JA 1h | 196.411 | 15.9985 | 0.925844 | 0.0557353 |
| Control 6h | 208.432 | 67.2275 | 0.693828 | 0.242889 |
| ABA 6h | 112.515 | 42.5103 | 0.358099 | 0.11753 |
| ACC 6h | 317.993 | 18.8565 | 1.07113 | 0.178352 |
| BABA 6h | 325.469 | 24.3705 | 1.12009 | 0.0660272 |
| Chitin 6h | 187.049 | 11.4662 | 0.678766 | 0.050546 |
| Epi 6h | 235.287 | 40.9841 | 0.780257 | 0.106099 |
| SA 6h | 202.789 | 49.9259 | 0.748219 | 0.121698 |
| Me-JA 6h | 214.315 | 84.3023 | 0.714432 | 0.237176 |
Source Transcript PGSC0003DMT400062333 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G35370.1 | +1 | 2e-76 | 253 | 133/299 (44%) | S-locus lectin protein kinase family protein | chr5:13588564-13591182 REVERSE LENGTH=872 |
