Probe CUST_52816_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52816_PI426222305 | JHI_St_60k_v1 | DMT400081499 | TGTCAAAAAAGAAGAAGGAAATATCCCAATTCATATCGACTCTCACACCTTCCGCTTTAC |
All Microarray Probes Designed to Gene DMG400031885
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52816_PI426222305 | JHI_St_60k_v1 | DMT400081499 | TGTCAAAAAAGAAGAAGGAAATATCCCAATTCATATCGACTCTCACACCTTCCGCTTTAC |
Microarray Signals from CUST_52816_PI426222305
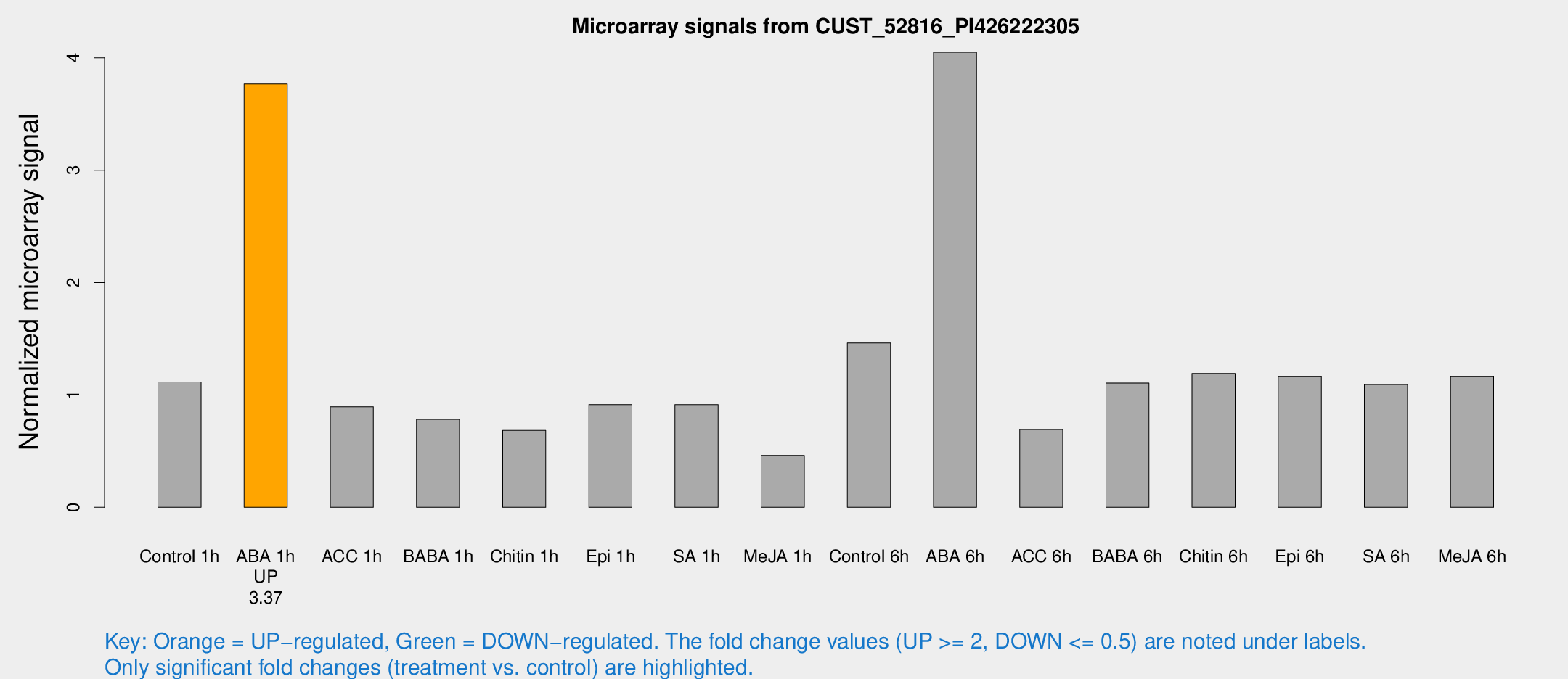
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 71.6797 | 9.47337 | 1.11625 | 0.0829616 |
| ABA 1h | 213.976 | 28.8913 | 3.76713 | 0.4921 |
| ACC 1h | 58.511 | 5.37846 | 0.895385 | 0.0752723 |
| BABA 1h | 50.0937 | 10.3893 | 0.783597 | 0.103015 |
| Chitin 1h | 38.8364 | 3.92194 | 0.684162 | 0.0689108 |
| Epi 1h | 51.7159 | 9.06166 | 0.914286 | 0.171285 |
| SA 1h | 59.5211 | 4.6982 | 0.913332 | 0.0896735 |
| Me-JA 1h | 25.6296 | 6.68422 | 0.462722 | 0.161662 |
| Control 6h | 94.7021 | 14.4468 | 1.46306 | 0.146712 |
| ABA 6h | 280.37 | 46.2654 | 4.04996 | 0.579316 |
| ACC 6h | 50.6823 | 5.49714 | 0.692453 | 0.166623 |
| BABA 6h | 80.3911 | 14.0669 | 1.1064 | 0.174887 |
| Chitin 6h | 80.3666 | 5.89187 | 1.19154 | 0.11268 |
| Epi 6h | 83.3685 | 7.41314 | 1.16327 | 0.12905 |
| SA 6h | 76.2535 | 22.105 | 1.09344 | 0.563293 |
| Me-JA 6h | 76.1271 | 16.1296 | 1.16298 | 0.155597 |
Source Transcript PGSC0003DMT400081499 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G62980.1 | +3 | 1e-13 | 51 | 25/45 (56%) | F-box/RNI-like superfamily protein | chr3:23273479-23276181 REVERSE LENGTH=594 |
