Probe CUST_52779_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52779_PI426222305 | JHI_St_60k_v1 | DMT400082693 | CTGCTGCAGCTTCTGGTTTTTTGAATGAAGCTTTGTATAGTTTTACTTGTATCTCTATAC |
All Microarray Probes Designed to Gene DMG400032766
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52779_PI426222305 | JHI_St_60k_v1 | DMT400082693 | CTGCTGCAGCTTCTGGTTTTTTGAATGAAGCTTTGTATAGTTTTACTTGTATCTCTATAC |
Microarray Signals from CUST_52779_PI426222305
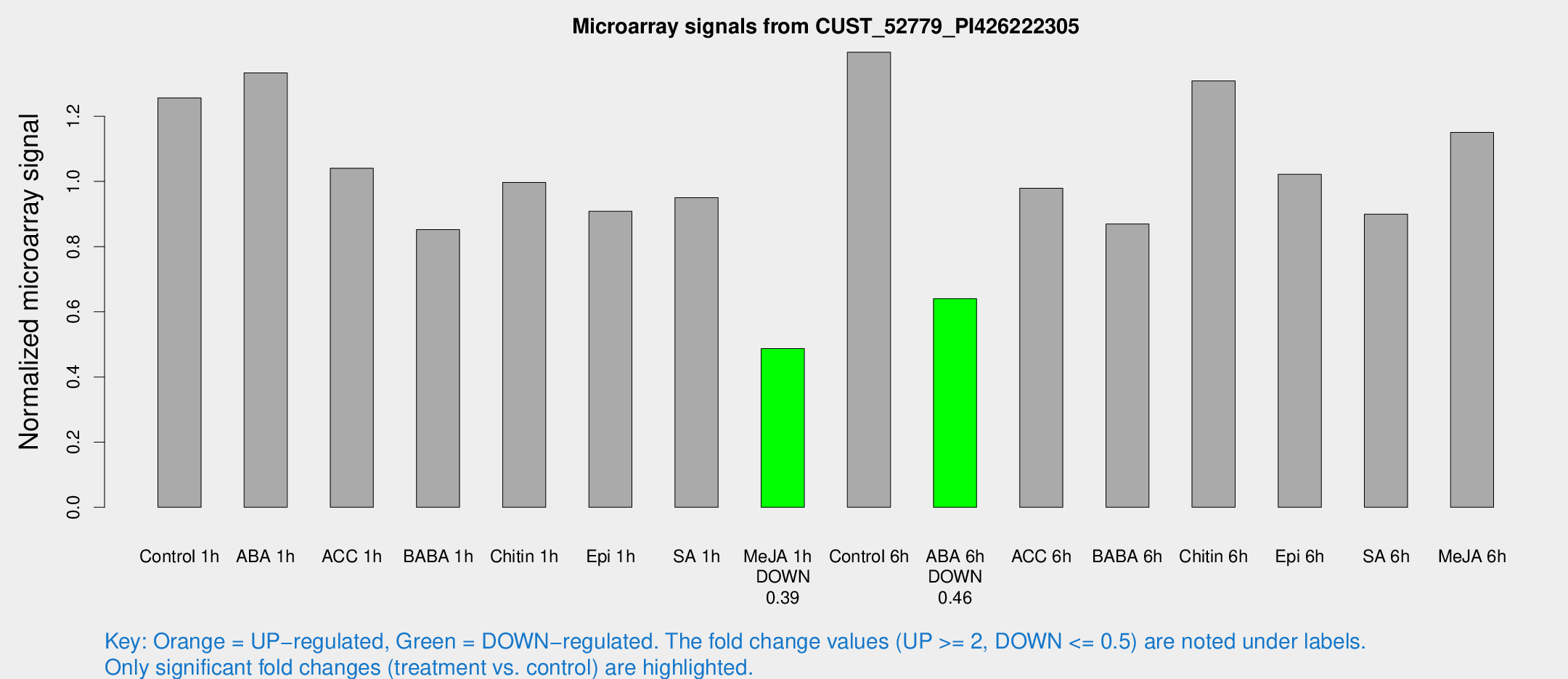
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1762.22 | 120.855 | 1.25615 | 0.0725689 |
| ABA 1h | 1654.56 | 118.531 | 1.33318 | 0.185596 |
| ACC 1h | 1550.9 | 282.501 | 1.04035 | 0.131653 |
| BABA 1h | 1157.41 | 112.316 | 0.852004 | 0.0492762 |
| Chitin 1h | 1251.57 | 72.3816 | 0.996625 | 0.0604091 |
| Epi 1h | 1097.31 | 63.4548 | 0.908095 | 0.0525109 |
| SA 1h | 1411.64 | 245.723 | 0.949952 | 0.141135 |
| Me-JA 1h | 557.497 | 35.7217 | 0.487215 | 0.0283187 |
| Control 6h | 2004.84 | 315.635 | 1.3964 | 0.104951 |
| ABA 6h | 961.534 | 105.367 | 0.640354 | 0.0370698 |
| ACC 6h | 1590.61 | 198.24 | 0.978781 | 0.0565768 |
| BABA 6h | 1397.04 | 242.902 | 0.869304 | 0.146026 |
| Chitin 6h | 1946.85 | 112.598 | 1.30872 | 0.0756147 |
| Epi 6h | 1635.83 | 203.524 | 1.02183 | 0.216089 |
| SA 6h | 1284.91 | 223.742 | 0.899607 | 0.0700767 |
| Me-JA 6h | 1688.26 | 382.812 | 1.15032 | 0.176108 |
Source Transcript PGSC0003DMT400082693 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G10560.1 | +1 | 1e-60 | 207 | 112/249 (45%) | plant U-box 18 | chr1:3484613-3486706 FORWARD LENGTH=697 |
